| Posted: Jul 01, 2014 |
Using fluorescent molecular rotors as a new tool to study critical protein interaction
|
|
(Nanowerk News) A recent study by scientists from the Agency for Science, Technology and Research (A*STAR) is the first to report on the use of fluorescent molecular rotors for cancer drug development. The study was published on 30 April as the cover article in the Journal of The American Chemical Society ("Molecular Rotors As Conditionally Fluorescent Labels for Rapid Detection of Biomolecular Interactions").
|
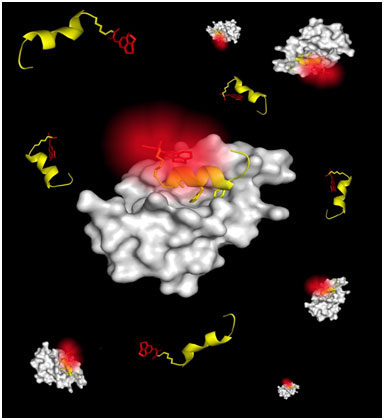 |
| The image shows molecular rotor 9-(2-carboxy-2-cyanovinyl) julolidine (CCVJ) molecules (in red) fluorescing after binding to MDM2 protein (in white). The image was featured on the front cover of the April issue of the Journal of the American Chemical Society. (Image: A*STAR)
|
|
On the cellular level, cancer is the uncontrolled growth of cells containing damaged or mutated DNA which could result in tumours. p53 is a ‘tumour suppressor protein’ because it functions as the body’s defence against cancer by binding to regulatory sites on the genome and trigger repair mechanisms to the DNA. Alternatively, p53 is also known to initiate a process of apoptosis or programmed cell death.
|
|
MDM2 protein is a negative regulator of p53. This means that a high level of MDM2 inhibits the activity of p53 by binding to it and breaking it down; on the other hand, when mutated or damaged DNA is detected, the level of MDM2 falls and allows p53 to initiate DNA repair. The breakdown of the regulatory abilities of MDM2 and p53 will lead to tumours. For instance, an overexpression of MDM2 has been observed in soft tissue sarcomas, gliomas, lymphomas and breast cancer. The study of MDM2-p53 interaction is, therefore, important in cancer research.
|
|
In this study, A*STAR scientists used fluorescent molecular rotors to study protein-protein interactions involving p53 and MDM2 in cells. They found that the fluorescent molecular rotor fluoresces or “lights up” when it is coupled with a short peptide fragment of the MDM2.
|
|
Armed with this finding, the scientists screened a library of small molecule fragments for candidates that may potentially disrupt p53-MDM2 binding. They detected a total of 15 hits – eight were validated by an existing method known as fluorescence polarisation and found extra seven which were missed out.
|
|
Dr Teo Yin Nah, Research Fellow at A*STAR’s Molecular Engineering Laboratory, said: “Researchers have used molecular rotors as viscosity sensor probes in live cells. This is the first time we have proved that molecular rotors can be used in a different way to understanding molecular interactions that causes cancer. Scientists now have another tool in their arsenal to further our understanding of the MDM2-p53 interaction.”
|

