| Posted: Feb 12, 2016 |
New nanotechnology detects biomarkers of cancer
(Nanowerk News) Researchers at Wake Forest Baptist Medical Center have developed a new technology to detect disease biomarkers in the form of nucleic acids, the building blocks of all living organisms.
|
|
The proof-of-concept study is currently published online in the journal Nano Letters ("Sequence-Specific Recognition of MicroRNAs and Other Short Nucleic Acids with Solid-State Nanopores").
|
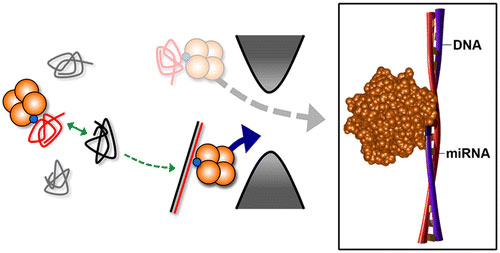 |
| The hybridization of a target nucleic acid with a synthetic probe molecule enables discrimination between duplex and single-stranded molecules with high efficacy. (© ACS)
|
|
"We envision this as a potential first-line, noninvasive diagnostic to detect anything from cancer to the Ebola virus," said Adam R. Hall, Ph.D., assistant professor of biomedical engineering at Wake Forest Baptist and lead author of the study. "Although we are certainly at the early stages of the technology, eventually we could perform the test using a few drops of blood from a simple finger prick."
|
|
Nucleic acids consist of chains or sequences of bases stretching from just a few to millions of elements long. The exact order in which these bases are found, even over short distances, is strongly tied to their functions, and therefore can be used as direct indicators of what is going on inside cells and tissue. For example, one family of these nucleic acids known as microRNAs are only about 20 bases long, but can signal a wide range of diseases, including cancer.
|
|
"Scientists have studied microRNA biomarkers for years, but one problem has been accurate detection because they are so short, many technologies have real difficulty identifying them," Hall said.
|
|
In the new technique, nanotechnology is used to determine whether a specific target nucleic acid sequence exists within a mixture, and to quantify it if it does through a simple electronic signature. "If the sequence you are looking for is there, it forms a double helix with a probe we provide and you see a clear signal. If the sequence isn't there, then there isn't any signal," Hall said. "By simply counting the number of signals, you can determine how much of the target is around."
|
|
In this study, the team first demonstrated that the technology could effectively identify a specific sequence among a background of competing nucleic acids, and then applied their technique to one particular microRNA (mi-R155) known to indicate lung cancer in humans. They showed that the approach could resolve the minute amount of microRNAs that can be found in patients. Next steps will involve expanding the technology to study clinical samples of blood, tissue or urine.
|
|
Hall holds a provisional patent on this technology.
|

