| Posted: July 9, 2009 |
DNA sorts out carbon nanotubes |
|
(Nanowerk News) DNA could be the answer to sorting different kinds of carbon nanotubes, say US researchers.
|
|
Short strings of synthetic DNA wind onto nanotubes in a very sequence-dependent way, which has allowed researchers to separate 12 of the most common types of tubes from the inevitable mixtures that form when nanotubes are synthesised. Since the physical and electronic properties of nanotubes are heavily dependent on the size and structure of the tube, separating the mixtures could lead to much more effective nanotube-based devices and even ways of making specific types of tubes selectively.
|
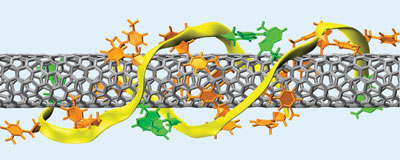 |
| Each DNA sequence wraps around a particular type of nanotube. (Image: Nature)
|
|
Ming Zheng of Dupont Central R&D, Delaware, and Anand Jagota from Lehigh University in Bethlehem, US, developed the technique after discovering a few years ago that DNA could wrap onto carbon nanotubes to form barrel-like structures. The real breakthrough came when they found that certain short sequences of DNA could selectively coil around tubes of a specific size and structure, allowing them to be fished out of mixtures of different tubes. 'This is a dramatic improvement in nanotube sorting,' says Zheng, 'if you look at all the methods available, none of them can achieve this level of separation. People could get one or two [different structures] from a mixture, but we have shown that we can get each and every major species out in a systematic way.'
|
|
The findings have been published in ("DNA sequence motifs for structure-specific recognition and separation of carbon nanotubes").
|
|
Single walled carbon nanotubes (SWNTs) come in three different varieties, depending on their structure. If you consider a flat sheet of graphitic carbon, or graphene, it could be rolled up in two perpendicular directions with the edges matching up perfectly. This gives rise to 'armchair' and 'zig-zag' type tubes (depending on which way you think of rolling up the sheet), which are generally metallic. However, if the sheet is rolled such that when the two edges meet to close the tube they are offset slightly, then the tube becomes 'chiral' due to its helical structure. Chiral SWNTs are usually semiconducting, but their electronic properties depend on the diameter of the tube and the degree of offset or chirality.
|
 |
| The DNA forms a barrel shape around the nanotubes according to their size and chirality (Image: Nature)
|
|
The DNA strands that the team use tend to be 8-30 bases long and contain repeated strings of two to four bases, normally containing a single purine base (A or G) and one or more pyrimidines (C and T). Unfortunately, explains Zheng, there doesn't seem to be any predictable correlation between the sequence and the selectivity with which it binds to nanotubes, and many of the sequences they tested did not give any separation. 'Mechanistically, we don't have a clear picture yet,' he adds, 'but different DNA sequences will have slightly different wrapping structures, and that structure is also dependent on the nanotube structure - the orientation or chirality of the tube, so the two have to match up somehow, but exactly what we mean by 'matching up' is something we don't really understand yet.'
|
|
Ralph Krupke, an expert in sorting and applications of carbon nanotubes from the Institute of Nanotechnology in Karlsruhe, Germany, is very impressed by the technique. 'It's a very interesting approach, because the interaction between the DNA and the nanotube is quite weak, especially compared to the polymers that people have used before to try separating tubes by chiral angle. It's virtually impossible to remove those polymers from the nanotube surface, which is a problem for making devices afterwards. With DNA, you can get rid of it relatively easily with a thermal treatment, so I think this is a technique which a lot of people will try out.'
|
|
|
|
However, Zheng points out that simply sorting out mixtures of nanotubes is not a complete solution to the problem, especially since the technique is not really amenable to large scale reproduction. 'It has to be coupled with a synthetic method,' he adds, 'and I'm really excited about the field of nanotube cloning - using a seed to grow identical, single chirality nanotubes - it's at a very early stage, but I think eventually purification and synthesis can join forces to solve the problem of nanotube polydispersity.'
|
|
Krupke agrees that the method would be ideal to produce seed tubes for cloning, particularly because separating the tubes involves sonication, which leads to shorter tubes that are less useful in device manufacture. 'The growth rates continuing from existing tubes are also rather low compared to growth rates out of catalysts,' he adds. 'The ultimate aim is to have chirality-specific growth of nanotubes [on catalysts], but that doesn't seem to be in reach yet, so right now this is state-of-the-art.'
|


