| Posted: September 8, 2009 |
Breakthrough in direct-method research advances detection of protein crystallographic structure |
|
(Nanowerk News) Recent years have witnessed advances in direct-method research on protein crystallography obtained by the research team led by Academician Fan Haifu from the Institute of Physics, Chinese Academy of Sciences (CAS). The direct-method Double-space Single-wavelength Anomalous Diffraction (SAD) Iterative Phasing Approach was proposed by the research team in 2004. Two years later, they put forward the new direct-method Double-space Molecular Replacement (MR) Iterative Model Building Approach. Both of them are remarkably more efficient compared with the original SAD and MR methods, as is shown in Figure 1 and Figure 2.
|
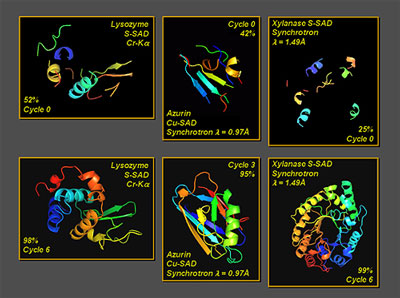 |
| Figure 1: The direct-method Double-space SAD Iterative Phasing Approach [Acta Cryst. (2004) D60, 1991-1996]
|
|
|
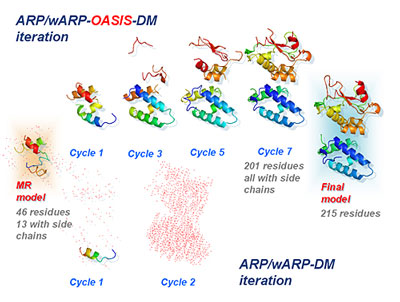 |
| Figure 2: The direct-method Double-space MR Iterative Model Building Approach [Acta Cryst. (2007) D63, 793-799]
|
|
CAS Institute of Physics has been working on the detection of protein crystallographic structure by combining the SAD method with the Single Isomorphous Replacement (SIR) method Since 1965,. This approach was considered to be the cynosure of international interests in the field of crystallographic research based on direct methods during the 1980's and 1990's. Most of the major research institutes across the globe engaged in direct methods took part in this attractive research, including two research groups led by Nobel laureates,. During 1984 and 1985, major improvements were obtained on the research approach originally proposed in 1965 by the Research Team on Methods for the Analysis of Crystallographic Structure at CAS Institute of Physics. Ever since then, the team's achievement in combining direct methods with SAD and SIR methods has generated positive influence on the research internationally.
|
|
Since the release of the computer program OASIS, successful analyses of some protein structures that pose tough problems for other programs have been conducted in China, United Kingdom of Britain, the U.S.A., France, Japan, Germany, etc. From 2005 to 2008, Prof. Fan and his colleagues were consecutively invited to important international symposia and seminars for 6 times to present introductions to the principle and operation of the program OASIS, which has always been undergoing updating by far. New version of OASIS with remarkably improved efficiency and automatic features is expected to be unveiled by the research team by the end of 2009.
|
|
|
|
The research was supported by the National Natural Science Fund of China (Award No.: 19674071). During the implementation of the project, the research team, for the first time in the world, succeeded in interpreting the formerly unknown protein crystallographic structure from a set of SAD data at 2Å resolution on the one hand, and in programming the first version of OASIS on the other hand.
|


