| Posted: October 14, 2009 |
NHGRI grant for nanopore technology to analyze DNA |
|
(Nanowerk News) The National Human Genome Research Institute (NHGRI) has awarded a $1.1 million grant to researchers in the Jack Baskin School of Engineering at UC Santa Cruz to support their work on nanopore technology for analyzing DNA.
|
|
Led by biomolecular engineers Mark Akeson and David Deamer, the UCSC nanopore group has pioneered a technology based on a tiny pore in a membrane, called a "nanopore" because it is just 1.5 nanometers wide at its narrowest point. The nanopore is formed by a self-assembling protein complex called an ion channel and is just big enough to allow a single strand of DNA to pass through. Researchers use the nanopore device to obtain precise measurements of DNA structure and dynamics as the molecule passes through the pore.
|
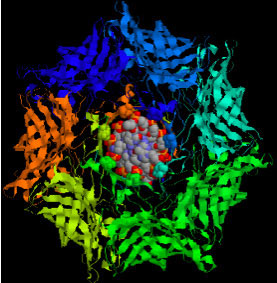 |
| This image of the nanopore used by UCSC researchers shows a double-stranded DNA molecule superimposed on the channel of the nanopore. The DNA molecule, 2.2 nanometers in diameter, fits into the pore vestibule but not through the pore, which is 1.5 nanometers.
|
|
A primary goal of the project is to develop nanopore technology as a fast and inexpensive method for DNA sequencing. Medical diagnosis and treatment is being revolutionized by tools that enable doctors to quickly obtain detailed genetic information about their patients. That genetic information is encoded in the sequence of nucleotide subunits in DNA molecules. Despite many advances in sequencing technology, however, DNA sequencing is still too expensive and time-consuming for routine clinical use.
|
|
Akeson, a professor of biomolecular engineering, said the UCSC nanopore group has made progress recently by coupling DNA-binding enzymes to the nanopore. DNA polymerases are enzymes involved in the replication of DNA in cells. When coupled to the nanopore, the enzymes control the movement of the DNA molecule through the pore.
|
|
"We are borrowing from nature, which has developed this molecular machinery to replicate DNA in cells," Akeson said. "The polymerase controls the rate at which the DNA is processed through the nanopore sensor, operating in the range of 1 to 100 milliseconds per nucleotide. It also regulates the distance the DNA molecule moves, so that it advances one nucleotide at a time."
|
|
In the work funded by the NHGRI grant, the researchers are focusing on experiments to measure the effects of voltage and other variables on how efficiently the nanopore system can control and process long DNA molecules (up to 2,500 nucleotides in length). The new grant was funded through the economic stimulus bill (the American Recovery and Reinvestment Act).
|
|
Since its beginnings in 1996, the UCSC nanopore project has grown into a large collaborative effort within the Baskin School of Engineering. In addition to Akeson and Deamer, a research professor of biomolecular engineering, the group now includes William Dunbar, assistant professor of computer engineering; Hongyun Wang, professor of applied math and statistics; and senior investigators Kate Lieberman, Felix Olasagasti, and Robin Abu-Shumays. Graduate students Noah Wilson, Daniel Garalde, and Nick Hurt are also associated with the nanopore group, as are six undergraduates.
|
|
|
|
"Some of the most promising work we do is coming from the undergrads in our lab," Akeson said. "One of our laboratories and four of our state-of-the-art nanopore devices are currently devoted to experiments by these students."
|
|
The nanopore technology developed at UCSC has been licensed by Oxford Nanopore Technologies of Oxford, U.K., which is developing nanopore technology for DNA sequencing and other potential applications. The UCSC Office for Management of Intellectual Property was instrumental in negotiating a favorable agreement with the company, Akeson said.
|

