| Posted: May 13, 2010 |
Graphene-DNA biosensor selective, simple to create |
|
(Nanowerk News) Graphene and DNA can combine to create a stable and accurate biosensor, reports a study published in the nanotechnology journal Small ("Constraint of DNA on Functionalized Graphene Improves its Biostability and Specificity"). The tiny biosensor might eventually help doctors and researchers better understand and diagnose disease.
|
|
Scientists at the Department of Energy's Pacific Northwest National Laboratory and Princeton University showed that single-stranded DNA strongly interacts with graphene, a nanomaterial made of sheets of carbon atoms just a single atom thick. They also found that graphene protects DNA from being broken down by enzymes similar to those found in body fluids - a characteristic that should make graphene-DNA biosensors highly durable.
|
|
"Graphene is of great interest because it has several unique characteristics, including being easy and relatively inexpensive to make," said PNNL chemist Yuehe Lin, the paper's corresponding author. "But very few had systematically explored how graphene interacted with DNA using multiple spectroscopic techniques until we took a look. We found they make quite the pair."
|
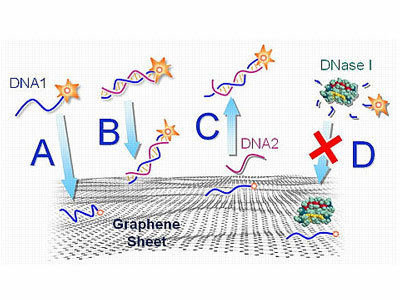 |
| An illustration of how fluorescent-tagged DNA interacts with functionalized graphene. Both single-stranded DNA (A) and double-stranded DNA (B) are adsorbed onto a graphene surface, but the interaction is stronger with ssDNA, causing the fluorescence on the ssDNA to darken more. C) A complimentary DNA nears the ssDNA and causes the adsorbed ssDNA to detach from the graphene surface. D) DNA adsorbed onto graphene is protected from being broken down by enzymes.
|
|
Scientists have been exploring the potential of nanotechnology - or tiny materials that are just one billionth of a meter in size - for several decades. A growing number of scientists are focusing on graphene because it is superconductive, is exceptionally strong and has a large surface area. It's also easier to make and use than other nanomaterials, such as carbon nanotubes. Nanotechnology could help create new drugs, deliver medicine and develop disease-detecting biosensors.
|
|
A graphene-DNA biosensor would detect diseases by fishing for molecules involved in disease. Like stringing a worm on a hook, scientists would place DNA from a gene that's known to contribute to a disease's development onto a piece of graphene. The researchers would then dip the biosensor hook into treated blood, saliva or another bodily fluid. If DNA from the disease-causing gene is in the fluid and takes the bait, the biosensor gives off a signal that scientists can detect.
|
|
The double-stranded nature of the DNA in our genes makes this fishing scheme possible. Normal double-stranded DNA looks like a twisted ladder. But single-stranded DNA looks like a comb: it's made up of a sequence of DNA letters, or bases, that stick up from the backbone and that look for another base to pair up with. When complementary sequences on single-stranded DNA meet, the bases form the rungs of the twisted ladder.
|
|
To design DNA-graphene biosensors, scientists need to understand how DNA and graphene interact. Lin and colleagues, including lead author and then-PNNL post-doctoral researcher Zhiwen Tang, attached a fluorescent molecule to DNA that glows when DNA floats freely to follow the DNA in test tubes. Next, they mixed the glowing DNA and graphene. Single-stranded DNA dimmed when it came in contact with graphene. But the brightness of double-stranded DNA decreased only slightly under the same conditions. Further analysis with several spectroscopy tests showed that graphene's interaction with single-stranded DNA is much stronger than with its double-stranded cousin. The tests also suggested that graphene altered single-stranded DNA's structure.
|
|
To find out if single-stranded DNA could be coaxed off the graphene by making it double-stranded, the researchers added plain, single-stranded DNA that had a complementary sequence of DNA bases. The original single-stranded DNA shined anew. This indicated the original single strand of DNA had combined with the added DNA strand and formed a new molecule that detached from graphene's surface.
|
|
The scientists then tested how picky the single-stranded DNA on the graphene was about partners. They placed the graphene-DNA biosensors into two different test tubes. In one, they added a complementary DNA strand with bases that were a perfect match to the DNA already attached to the graphene. In the other, they placed a complementary DNA strand that had one base that didn't pair up with the original DNA strand on the graphene surface.
|
|
Both gave off more light after the complementary DNA was introduced. But light from the tube with the perfectly matched DNA strands was two times brighter than from the tube with the slightly mismatched DNA strands. The ability to identify whether a target DNA strand has been found within one base match - called high specificity - should make graphene-DNA biosensors more accurate than other, conventional linear DNA biosensors, the scientists wrote.
|
|
Graphene also helps make DNA durable, the scientists learned. They placed two kinds of single-stranded DNA - one that was attached to graphene, and another that was free floating - in test tubes. They added DNAse - an enzyme that chews up DNA - to both and found that the free DNA strands were broken down, while the graphene-DNA nanostructures remained intact for at least 60 minutes. The scientists suggested this protection could make DNA-graphene platforms that are well suited for imaging and gene delivery in patients.
|
|
"The simple design and tremendous durability of graphene-DNA biosensors make diagnosing life-threatening diseases with them a possibility," Lin said. "Now my colleagues and I will look to see if graphene's ability to protect DNA against enzymes could help DNA-graphene structures deliver drugs to diseased cells or even help with in gene therapy."
|
|
Princeton University provided the graphene and PNNL's Transformational Materials Science Initiative paid for this study. Some of the research was conducted at EMSL, the Environmental Molecular Sciences Laboratory, a national scientific user facility located at PNNL.
|

