| Posted: May 10, 2007 |
DNA sieve: nanoscale pores can be tiny analysis labs |
|
(Nanowerk News) Imagine being able to rapidly identify tiny biological molecules such as DNA and toxins using a system that can fit on a microchip or in a drop of salt water. It’s closer than you might think, say a team of researchers from the National Institute of Standards and Technology (NIST), Brazil’s Universidade Federal de Pernambuco, and Wright State University in Dayton, Ohio.
|
|
In a paper appearing next week in the Proceedings of the National Academy of Sciences ("Single-molecule mass spectrometry in solution using a solitary nanopore" - open access article), the team proves for the first time that a single nanometer-scale pore in a thin membrane resembling a cell wall can be used to accurately detect and sort different-sized polymer chains (a model for biological molecules) that pass through the channel.
|
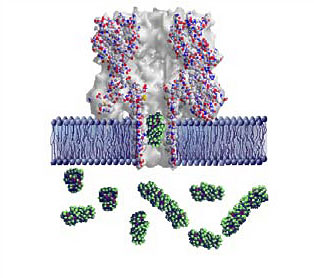 |
| Graphic showing a lipid bilayer membrane (blue) with an alpha-hemolysin nanopore. A polyethylene glycol molecule (green globular structure) is transiting the pore; others are in solution on one side of the membrane. The colored spheres represent individual atoms, and are approximately 0.5 nanometers in diameter, or one twenty-thousandth the width of a human hair. (Image: NIST)
|
|
Traditionally, unknown molecules in solution are measured and identified using mass spectrometry, a process that involves ionizing and disintegrating large numbers of the target molecule, then analyzing the masses of the resulting fragments to produce a “molecular fingerprint” for the original sample. The required equipment can cover a good-sized desk. By contrast, the “single-molecule mass spectrometry” system described in the PNAS paper is a non-destructive technique that in principle can measure one molecule at a time in a space small enough to fit on a single microchip-based device the size of a cell phone or PDA.
|
|
The technique involves creating a “lipid bilayer,” a membrane of biological molecules similar to the walls of living cells, and creating a pore in it with a protein such as alpha-hemolysin, produced by the Staphyloccoccus aureus bacteria specifically to poke holes in cells. The target molecules are forced one-at-a-time through the nanopore, which is only 1.5 to 12 nanometers in diameter, by an applied electric current. As the molecules—in this experiment, different-sized chains of the polymer polyethylene glycol (PEG)—pass through the channel, the current flow is reduced in proportion to the size of each individual chain, allowing an easy measure of its mass.
|
|
As a control, a solution of a highly purified PEG of a specific size was characterized with the nanopore. The resulting “fingerprint” closely matched the one identifying samples of the same size polymer in the mixed-chain solution.
|
|
Further enhancement of the data from both the experimental and control tests yielded mass measurements and identifications of the different PEG chains that correlate with those made by traditional mass spectrometry.
|
|
Because the dimensions of the lipid bilayer and the alpha-hemolysin pore, as well as the required amount of electrical current, are at the nanoscale level, the “single-molecule mass spectrometry” technology may one day be incorporated into “lab-on-a-chip” molecular analyzers and single-strand DNA sequencers.
|

