| Dec 10, 2010 |
Light games with DNA
|
|
(Nanowerk News) The diagnosis of hereditary diseases and the identification of genetic fingerprints hinge on high-sensitivity DNA imaging biotechnologies. These imaging tools detect specific genes in cells using fluorophores—fluorescent tags that can illuminate DNA structures—and quenchers that interact with these tags to prevent them from emitting light, effectively working as an 'off switch'.
|
|
In a development that expands the detection toolbox and the genetic alphabet, a team led by Ichiro Hirao from the RIKEN Systems and Structural Biology Center, Yokohama, has now designed an artificial base pair between a fluorophore (Dss) and quenchers (Pn and Px) ("A New Unnatural Base Pair System between Fluorophore and Quencher Base Analogues for Nucleic Acid-Based Imaging Technology"). This method incorporates the pair into complementary DNA strands using polymerases and demonstrates that either Pn or Px can decrease the fluorescence of Dss upon hybridization (Fig. 1).
|
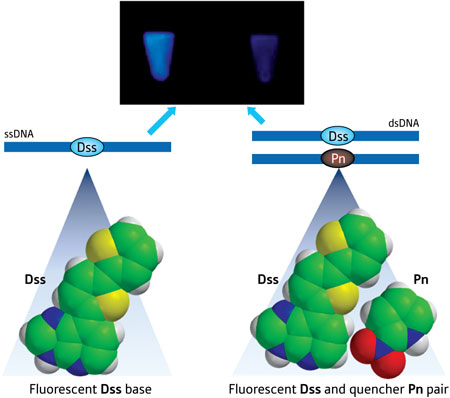 |
| Figure 1: Schematic illustration showing fluorescence of the Dss base (left) and the Pn base (right) quenching the fluorescence upon hybridization.
|
|
Hirao and his team previously developed artificial base pairs involving Dss because of its strong fluorescence, which could illuminate DNA and RNA structures. "This time, we can put out the candle lit by Dss using the quencher as its pairing partner at will," he says.
|
|
Hirao notes that this ability is unique because fluorescent dye Dss and quencher Pn face each other on their respective ssDNA strand, forming an artificial DNA base pair that also works in biological systems. He says that this close proximity results in strong 'contact quenching' of the fluorophore.
|
|
Usually, researchers have attached fluorophores and quenchers to natural bases through a linker that mediates so-called fluorescence resonance energy transfer (FRET) between dyes. However, this process lacks efficiency compared to contact quenching. Also, according to Hirao, unlike the Dss–Pn system, typical fluorophore–quencher pairs cannot be introduced at specific positions in DNA strands using polymerases, limiting their applications.
|
|
After establishing that the pairs were compatible with natural DNA synthesis techniques, Hirao's team integrated the Dss–Pn pair in the stem of molecular beacons—hairpin-shaped single-stranded DNA (ssDNA) structures that fluoresce upon hybridization with DNA targets. They found that the beacons detected the targets with high sensitivity and differentiated ssDNA containing one mismatched base.
|
|
Next, the researchers tested the performance of Dss–Px in polymerase chain reaction (PCR)—a powerful DNA amplification technique. Dss-bearing ssDNA fragments became less fluorescent upon assimilation of Px into synthesized DNA chains, allowing the team to monitor the amplification process in real time.
|
|
"One of our present tasks is to apply this system to in vivo cell experiments," says Hirao. "If it is possible, we will be able see the on–off of a specific gene expression."
|

