| Aug 31, 2011 |
New, highly sensitive microarray can detect 300 copies of microRNA in a one-microliter sample
|
|
(Nanowerk News) MicroRNAs are important regulators of gene activity, and their malfunction is known to be responsible for certain cancers and many other diseases. The rapid detection of microRNAs using small-volume samples could therefore help save millions of lives. Such a technique could also pave the way for widespread analysis of the spatial and temporal patterns of gene activity involving microRNAs, which are believed to play a critical role in the development and operation of organisms.
|
|
Somenath Roy at the A*STAR Institute of Bioengineering and Nanotechnology and co-workers have now developed a microfluidic-assisted technique for microRNA detection ("A microfluidic-assisted microarray for ultrasensitive detection of miRNA under an optical microscope "). The method is so sensitive that it can detect 300 copies of target microRNAs in a sample volume of just one microliter. More importantly, the technique is extremely efficient – it can produce results in around 30 minutes while conventional technologies can take up to 18 hours.
|
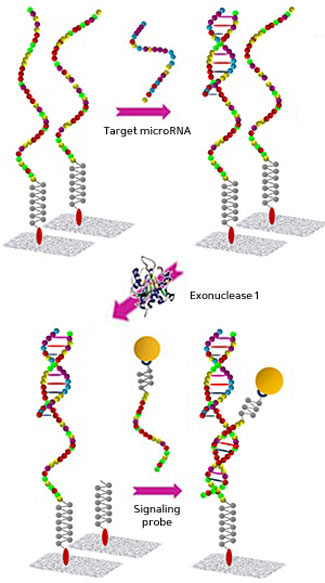 |
| The sequence of an ultrasensitive assay for detection of microRNAs on a chemically modified glass slide.
|
|
MicroRNAs, small chains of genetic material approximately 22 nucleotides long (genes typically contain thousands of nucleotides), control gene activity by attaching themselves to the gene transcripts used in the manufacture of proteins in cells. Once attached, they degrade the transcripts or otherwise interfere with their function. MicroRNAs have been found in all nucleated cells except in fungi, algae and marine plants. In an individual cell, there are generally hundreds, sometimes thousands, of different microRNAs.
|
|
Because they are so small, however, microRNAs are difficult to detect. Most techniques developed so far to identify and track microRNAs required the number of microRNAs in the sample to be amplified prior to detection. The new technique developed by Roy and his co-workers uses microarrays with probes to capture individual microRNA chains differing by just a single nucleotide without amplification.
|
|
The microRNAs are detected and marked with nanoparticles of gold (see image), which can then be counted directly by differential interference contrast microscopy, providing information on the amount of any particular form of microRNA chain. The whole process is accelerated by applying microfluidic convection flow to circulate the sample across the microarray.
|
|
The researchers tested their new system by analyzing the differences in activity between healthy and tumor cells of a family of microRNAs known as hsa-let-7. Their results were consistent with published data obtained by conventional techniques.
|
|
In the future, the researchers intend to develop a platform that would be able to identify microRNAs for diagnostic purposes directly from samples of body fluids, such as blood, saliva and breast milk.
|

