| Sep 30, 2011 |
New method for single-cell analysis may improve cancer treatment
|
|
(Nanowerk News) A collaboration between Danish and American researchers has resulted in the development of a new method that enables the measurement of enzyme activities in individual human cells ("Detection of Single Enzymatic Events in Rare or Single Cells Using Microfluidics"). This method can be used to measure how cell-to-cell variation in tumours affects the overall response to chemotherapy and thus clarify some of the molecular causes of the chemo-resistance often seen in cancer patients. In the long run, the researchers hope that the method can be used to target chemotherapy to those patients who benefit from this treatment.
|
|
The enzyme activities measured by this method belong to the enzyme group called DNA topoisomerases. The members of this enzyme family are important in the systemic treatment of a considerable number of cancers, where several of the commonly used chemotherapeutic agents act specifically by interfering with the activity of the topoisomerases. Unfortunately, it appears that many patients are either already resistant to topoisomerase-directed chemotherapy at the start of treatment or develop resistance after initial treatment. The reasons for this resistance are not fully determined, but several studies suggest that there is a direct connection between a cell's topoisomerase activity and the cytotoxic effect of chemotherapeutic agents.
|
|
Cancer cells are typically characterised by a high level of topoisomerase activity compared to the body's healthy cells, and this is why chemotherapy of this type affects tumours to a larger extent than normal body cells. Tumours are also characterised by a very high degree of cellular diversity, which means that the individual cells forming a tumour are very different. How this difference between cells affects the overall tumour response to chemotherapy is still unknown, but it is expected to be one of the reasons for the chemo-resistance seen in some patients.
|
|
The newly developed method makes it possible for the first time to examine the differences with respect to the topoisomerase activity between individual cells in a cancerous tumour and compare these with the tumour's chemo response, which could contribute significantly to the understanding of the molecular basis of chemo-resistance.
|
|
Description of the new method
|
|
The newly developed method for measuring topoisomerase activity in individual cells is illustrated in the figure. The method is based on a so-called microfluidic system combined with special DNA sensors that enable the detection of each catalytic reaction mediated by topoisomerase.
|
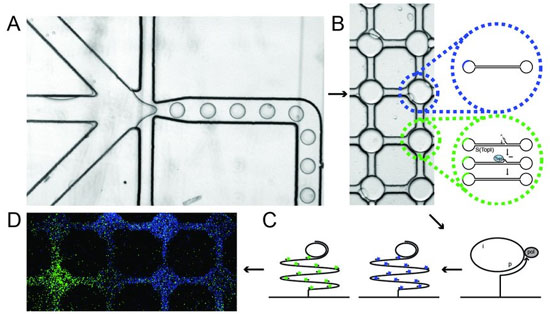 |
| Measurement of DNA topoisomerase activity. A. Cells, DNA sensors and a special buffer – which destroys the cell membrane and allows the cellular topoisomerase enzymes to react with DNA sensors – are loaded into each channel of the system. B. The reaction of the topoisomerases with the DNA sensor, which consists of a piece of self-folding DNA transformed into a closed DNA circle upon reaction with the topoisomerase. C. After this reaction, the mixture is transferred onto a glass plate that is already activated with so-called DNA primers. These support a so-called DNA amplification process resulting in products consisting of 103 copies of the circular DNA sensors that can be made visible at the single-molecule level through hybridisation to fluorescent probes. In addition to the described topoisomerase sensor, a pre-made DNA circle that serves as a control is added to the system. The circular topoisomerase sensors are visualised with green fluorescent probes, while the control circles are visualised with blue fluorescent probes. D. Example of the detection of topoisomerase activity (green signals) and control circles (blue signals) in the system. Each field displays the contents of a single lipid-encapsulated droplet. (Figure: Sissel Juul)
|
|
Part A of the figure shows how cells, DNA sensors and a special buffer – which destroys the cell membrane and allows the cellular topoisomerase enzymes to react with DNA sensors – are loaded into each channel of the system. These components are then mixed in very small lipid-encapsulated droplets, in which the topoisomerases react with the DNA sensor.
|
|
Part B shows the reaction of the topoisomerases with the DNA sensor, which consists of a piece of self-folding DNA transformed into a closed DNA circle when it reacts with the topoisomerases. After this reaction, the mixture is transferred onto a glass plate that is already activated with so-called DNA primers. These support a so-called DNA amplification process resulting in products consisting of 103 copies of the circular DNA sensors that can be made visible at the single-molecule level through hybridisation to fluorescent probes. In addition to the described topoisomerase sensor, a pre-made DNA circle that serves as a control is added to the system. The circular topoisomerase sensors are visualised with green fluorescent probes, while the control circles are visualised with blue fluorescent probes.
|
|
Part D of the figure shows an example of the detection of topoisomerase activity (green signals) and control circles (blue signals) in the system. Each field displays the contents of a single lipid-encapsulated droplet.
|
|
The next step
|
|
The researchers' new findings will form the basis for numerous measurements of cancer cells, which will hopefully lead to a greater understanding of the mechanisms underlying chemo-resistance. In the long run, a clarification of this resistance will help doctors target chemotherapy treatment to those patients who will benefit from it.
|
|
The video illustrates how the encapsulation of cells in droplets occurs when the system is running:
|
|
|

