| May 02, 2012 |
3D insights into the molecular teamwork in biomembranes
|
|
(Nanowerk News) For chemists, cellular biomembranes are hard nuts to crack. It is difficult to analyze proteins that are firmly anchored in biomembranes using standard biochemical methods; and it is even more difficult to investigate their three-dimensional structure and interaction with other proteins. A group of researchers led by Prof. Dr. Anne S. Ulrich at the Karlsruhe Institute of Technology (KIT) have developed a method that enables them to take a close look at individual atoms, and even at the atoms' natural environment in the lipid bilayer. How do antimicrobial peptides act on the membrane of a bacterium? How does a virus manage to penetrate the envelope of the host cell? How is the information from hormonal and neurotransmitter signals translated into the cell's interior? Can this knowledge be used to produce new substances whose molecular properties can be accurately defined, for example substances for the treatment of cancer and infections?
|
|
Around thirty per cent of all cellular proteins are in some way or other associated with a biomembrane – some proteins cross the lipid bilayer once and some even several times while others are bound to membranes and protrude into the extra- or intracellular space. The pharmaceutical industry has long known that cellular membranes are important drug targets. Aspirin acts on a specific membrane protein, thereby preventing the transmission of pain signals, to cite just one example. Researchers are currently particularly interested in the structure-function relationship of membrane proteins and membrane-active peptides. Their targets include ion channels, signal receptors, peptide antibiotics as well as Trojans that are used to shuttle drugs across the cell membrane.
|
|
"In all these cases, we are not only interested in the three-dimensional structure of these molecules; we are also interested in how these molecules interact with each other," said Prof. Dr. Anne S. Ulrich from the Institute of Biological Interfaces 2 (IBG-2) at the Karlsruhe Institute of Technology (KIT). "We are looking at the molecules when they bind to membranes; we specifically manipulate the molecular contact areas and try to deduce their mechanisms of action from this information."
|
|
A special trick
|
|
Molecules located on or in a membrane are difficult to analyze. Intractable forces prevail in lipid bilayers, which chemists who want to isolate individual molecules need to overcome. However, the three-dimensional structure of a membrane that is investigated in an aqueous solution provides little information about the membrane's structure in its natural environment where specific molecules are incorporated and interact with lipids and other proteins in a way that is difficult to predict. Crystallographic methods only provide good results for molecules that are removed from the lipid bilayer and are present as purified crystals.
|
|
"This is why around 15 years ago I started developing a different method that was specifically applicable to membrane-active peptides and membrane proteins," said Ulrich. "The optimized version of this method, which is known as solid-state nuclear magnetic resonance spectroscopy (NMR), enables us to look at the accurate three-dimensional structure of proteins in their natural membrane environment." The researchers use a special trick, namely fluorine substitutes, to do this. Fluorine is not found in biological tissue, which is why the NMR measurement cannot be falsified by background noise. The sensitivity of the method is extremely high and the excellent resolution enables the specific visualization of the labelled segments.
|
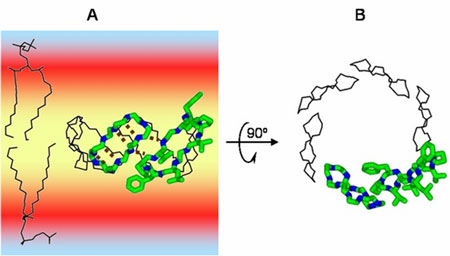 |
| Antimicrobial peptides (e.g. gramicidin S) form pores in bacterial membranes and ensure that the cellular content of the microorganisms flows out. (© Prof. Dr. Anne S. Ulrich)
|
|
Ulrich and her team now use this method routinely. If everything goes to plan, they are able to clarify the three-dimensional structure of a peptide molecule in a lipid bilayer within six weeks. Until recently, researchers used classical methods that took several years to obtain a result. The method is also very useful for analyzing the structure-function relationships of membrane-active peptides. For example, Ulrich's team managed to find out how antimicrobial peptides, which are found in the skin of frogs and in human sweat, attack the cell membrane of bacteria. In contrast to the majority of antibiotics developed by the pharmaceutical industry, the antimicrobial peptides tested by Ulrich and her group accumulate on the envelope of a pathogen and form a pore – the bacterium is perforated and its contents flow out. This mechanism targets the physical properties of the membrane, which prevents the bacterium from adapting to the altered situation by evolving mutations and resistance to the substance. This appears to be a promising new approach in the search for new antibiotics.
|
|
Using the new solid-state NMR method, Ulrich's team was able to clarify the molecular architecture of different peptides. This now enables them to explain how the molecules group together and form pores. The peptides are highly selective and do not attack the host's own cell membrane.
|
|
It is interesting to note that the chemical composition of some antimicrobial peptides is similar to that of so-called fusion peptides. Viruses that attack a host cell catapult these fusion peptides into the membrane of the host cell where they lead to the fusion of the two membrane envelopes. Comparative NMR measurements have shown that both types of peptides can induce similar mechanical disturbances in the lipid bilayer when they enter into unspecific interactions. However, nature has optimized the antimicrobial molecules in a way that does not normally happen; a few molecular "missiles" can induce the controlled formation of pores. A fusion machinery activates these weapons by way of a multi-tier cascade as they would otherwise stick to each other in an uncontrolled way. If these fusogenic peptides were given free rein they would form fibrils similar to those found in the tissue of Alzheimer's patients, and immediately destroy the membrane.
|
|
Signal transduction caught in the act
|
|
Another example from the field of signalling research shows that receptor proteins control the transmission of information between cells. These receptor proteins are anchored in the cellular membrane by way of one or several transmembrane segments. If an exogenous hormone docks to the cell membrane, two receptor proteins assemble in such a way that the regions that protrude into the cytosol are able to interact with cellular signalling molecules which then transfer the signal to the cell nucleus or to other molecular control centres. Which structural interactions now occur in the transmembrane regions of the receptor pair when a ligand has bound and the information is transmitted across the membrane?
|
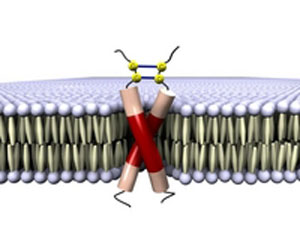 |
| A dimer consisting of E5 molecules binding a papillomavirus to the dimer of a membrane receptor which is normally activated by hormones. (© Prof. Dr. Anne S. Ulrich)
|
|
Ulrich and her team are currently investigating a receptor that besides being switched on by a hormone, is also switched on by the small papillomavirus protein E5. The interaction with E5 activates a signalling cascade that causes a cell to grow in an uncontrolled manner, like a cancer cell. "Crystallographic methods are not suitable for finding out which E5 and receptor areas interact with each other and what happens spatially," said Ulrich. Using a combination of different NMR methods, the researchers succeeded in clarifying the molecular composition of E5 and of the receptor in its membrane-bound state. Each is preferentially present as a dimer. The next step will be to catch the mixed proteins in the act, i.e. the point at which E5 binds to the membrane segments of the receptor, thus imitating the situation in which an exogenous hormone docks to the membrane.
|
|
In addition to solid-state NMR, Ulrich and her KIT group have recently installed a circular dichroism beamline in the KIT's ANKA synchrotron facility. This particle accelerator generates high energy radiation in the UV range, which is used for the qualitative clarification of molecular structures. This device leads to far better results than any other commercial laboratory device. "We use the device for very basic experiments in order to investigate whether and how a protein segment is folded in a membrane, or whether it has aggregated and has lost its function," said Ulrich. The subsequent solid-state NMR measurements of the same samples are highly accurate; individual atoms and even the atoms' movements can be visualized. Solid-state NMR is the method of choice for researchers who want to find out step by step what is actually happening in and around a membrane.
|


