| Posted: January 28, 2008 |
Molecular walker takes baby steps |
|
(Nanowerk News) Researchers at the California Institute of Technology report they are able to program the pathways by which DNA molecules self-assemble, and hence to engineer diverse dynamic functions at the molecular level.
|
|
"This capability is essential for something like the memory of a DNA computer, which would need large groups of molecules that can toggle from the on/off position in a fast and reliable fashion," said National Science Foundation (NSF) Program Manager Kathy Covert.
|
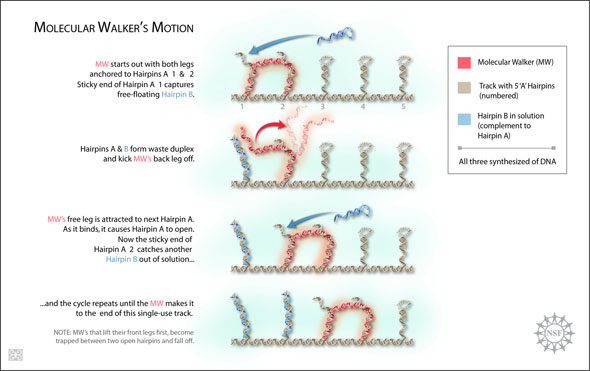 |
| Researchers have synthesized a molecular structure designed from single and double-stranded pieces of DNA that responds to a similarly modeled track, by walking down it autonomously. The walker places foot-over-foot as each appendage is attracted biochemically to the next hairpin along the track. As foot and hairpin make contact, the hairpin unravels. The free end of the hairpin then catches a complementary hairpin that is free-floating in the solution that the whole system is immersed in. Both hairpins coil together forming a "waste duplex" (or the familiarly shaped double helix), releasing the walker's foot for its next stride. If the walker reaches the end of the track successfully, it leaves behind non-reusable material, and the track is spent. Its travels are more perilous than may seem if it lifts the wrong foot and finds itself trapped between two open hairpins, the walker will fall off never seeing the end of its track. (Image: Zina Deretsky, National Science Foundation after figure by Peng Yin, Harry M. T. Choi, Colby R. Calvert and Niles A. Pierce, California Institute of Technology)
|
|
Researchers Peng Yin, Harry Choi, Colby Calvert and Niles Pierce, who are funded by NSF, report their research results in the Jan. 17 issue of Nature. To illustrate their approach for encoding self-assembly and disassembly pathways into DNA sequences, the researchers experimentally demonstrated the locomotion of a two-legged DNA walker that moves along a DNA track without human intervention.
|
|
Scientists are working to develop dynamic molecular systems for therapeutic, biosensing, and other applications. "Exploiting self-assembly is essential to constructing a molecule with the features we want it to have," Covert said.
|
|
This research was supported by NSF continuing grant #0533096 for collaborative research managed by the Chemical Bonding Center for Molecular Cybernetics sponsored by Columbia University. Led by Principal Investigator Milan Stojanovic, the center's goal is to produce synthetic molecular machines powered by molecular bond formation.
|
|
Luis Echegoyen, director of the NSF Division of Chemistry, said "this is one excellent example of the scientific outcome of synergistic activities developed within the CBC program. We are confident that similarly significant work derived from interdisciplinary research in CBCs will continue to emerge."
|

