| Posted: June 23, 2008 |
Automated microfluidic device reduces time to screen small organisms for genetic studies |
|
(Nanowerk News) Genetic studies on small organisms such as worms and flies can now be done more quickly using a new microfluidic device developed by engineers at the Georgia Institute of Technology.
|
|
The new "lab-on-a-chip" can automatically position, image, determine the phenotype of and sort small animals, such as the worm Caenorhabditis elegans that is commonly used for biological studies.
|
|
"Classical genetic approaches require altering genetic information and monitoring changes in a large number of animals, which can be excruciatingly slow and often requires manual manipulations," said Hang Lu, an assistant professor in Georgia Tech School of Chemical and Biomolecular Engineering. "As researchers move from studying single genes to analyzing interactions and networks, studies that require large sample sizes will be critical and this device allows for consistent and reliable operation to rapidly screen many animals."
|
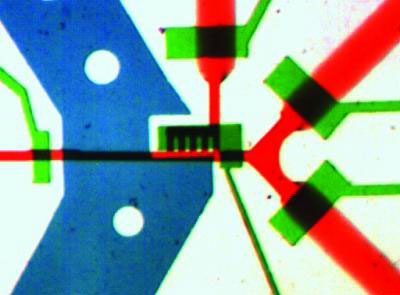 |
| An image of the microchip filled with dye to show the loading regulator (left green channel), temperature control (blue channel), detection zone (center black grates) and outlets (right red channels). The outlet channels are 100 microns wide. (Image: Image courtesy of Hang Lu)
|
|
In the July print issue of the journal Nature Methods, available online June 22, Lu and graduate students Kwanghun Chung and Matthew Crane describe their automated microsystem and initial experimental results. The results show that they can sort small organisms without human intervention based on cellular and subcellular features, or traits, with a high degree of accuracy at a rate of several hundred animals per hour. This work was funded by the National Science Foundation and the National Institutes of Health.
|
|
Using the microfluidic system is simple. Each small animal is automatically loaded into the microchip. The setup automatically arranges each organism in an identical position in the chip to reduce the processing time and increase throughput.
|
|
Once the organism is loaded, it is briefly immobilized by an integrated local temperature control system that cools the animal to approximate four degrees Celsius. Cooling effectively stops the animal's motion and allows repeated imaging of the same organisms because unlike commonly-used anesthetic drugs, the cooling doesn't have long-term effects.
|
|
After cooling, the system uses a high-resolution microscope to acquire multi-dimensional images of the animal on-chip.
|
|
"The advantage of using our microchip is that it's completely compatible with any standard microscope you'd find in a biology laboratory – epifluorescence, stereo, multi-photon or confocal – with no modification required," explained Lu.
|
|
The researchers have shown that the intensity and patterns of fluorescent markers imaged inside cooled animals versus those in anesthetized animals exhibit no discernible differences. Based on each animal's phenotype, or how each animal looks under the microscope, the computer identifies whether it is wild-type or mutant and sorts it into the appropriate group.
|
|
Initial tests to assess the system were conducted on C. elegans, one of the tiniest multi-cellular organisms that share many fundamental cellular/molecular mechanisms with more advanced organisms. However, the automated system can also be adapted to study other small organisms such as fruit flies and fish embryos.
|
|
For one experiment, Lu and her team tested the ability of the system to analyze the gene expression pattern – the intensity, location and timing of appearance of a fluorescent protein – in a population of organisms. They were able to sort the free-moving animals into two categories, those fluorescing in a particle neuron and those that are not, at a speed of approximately 900 animals per hour. More than 90 percent of the animals were loaded into the observation chamber within 0.3 seconds after the previous animal exited.
|
|
In another experiment, the researchers were successful in separating a small number of mutant animals from a large population of wild-type animals based on the fluorescence in a single pair of neurons. With on-line processing and decision-making without human supervision, the system achieved a sorting speed of approximately 150 animals per hour and a false negative rate of less than 0.2 percent, indicating that almost all the mutants were captured by the system.
|
|
A third experiment was aimed at demonstrating the ability of the system to screen organisms based on micro-sized synaptic features of the animals. Results showed that the system was able to sort mixed populations at a rate of approximately 400 animals per hour for this application. In all three experiments, it would have taken researchers much longer to identify the worms manually with high-resolution microscopy a few worms at a time.
|
|
"This is the first automated device to combine high-resolution imaging with automated sorting of the worms." added Lu. "Now that we have the automated system, we are able to perform genetic screens a lot faster than what has traditionally been done and speed up the discovery of new genes, new functions and new pathways."
|

