| Oct 27, 2014 |
Imaging the genome: cataloguing the fundamental processes of life
|
|
(Nanowerk News) A new study at the University of Cambridge has allowed researchers to peer into unexplored regions of the genome and understand for the first time the role played by more than 250 genes key to cell growth and development.
|
|
The team of researchers, led by Dr Rafael Carazo Salas from the Department of Genetics, combined high-resolution 3D confocal microscopy and computer-automated analysis of the images to survey the fission yeast genome with respect to three key cellular processes simultaneously: cell shape, microtubule organisation and cell cycle progression. Microtubules are small, tube-like structures which help cells divide and give them their structure.
|
|
Of the 262 genes whose functions the team report in a study published today in the journal Developmental Cell ("A Genomic Multiprocess Survey of Machineries that Control and Link Cell Shape, Microtubule Organization, and Cell-Cycle Progression"), two-thirds are linked to these processes for the first time and a third are implicated in multiple processes.
|
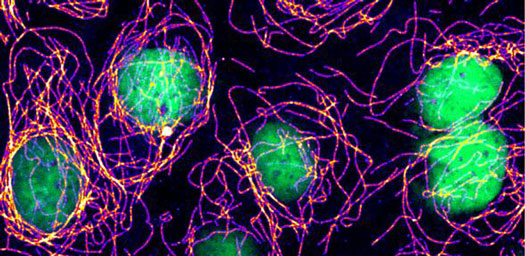 |
| Cells with damage in their DNA (green) assemble abnormally stable microtubule structures (purple to white). This new link between microtubule control and the response to DNA damage, originally discovered in yeast, can be observed also in human cells. (Image: L. Wagstaff, E. Piddini)
|
|
“More than ten years since the publication of the human genome, the so-called ‘Book of Life’, we still have no direct evidence of the function played by half the genes across all species whose genomes have been sequenced,” explains Dr Carazo Salas. “We have no ‘catalogue’ of genes involved in cellular processes and their functions, yet these processes are fundamental to life. Understanding them better could eventually open up new avenues of research for medicines which target these processes, such as chemotherapy drugs.”
|
|
Using a multi-disciplinary strategy that took the team over four years to develop, the researchers were able to manipulate a single gene at a time in the fission yeast genome and see simultaneously how this affected the three cellular processes. Fission yeast is used as a model organism as it is a unicellular organism – in other words, it consists of just one cell – whereas most organisms are multicellular, yet many of its most fundamental genes carry out the same function in humans, for example in cell development.
|
|
The technique enabled the researchers not only to identify the functions of hundreds of genes across the genome, but also, for the first time, to systematically ask how the processes might be linked. For example, they found in the yeast – and, importantly, validated in human cells – a previously unknown link between control of microtubule stability and the machinery that repairs damage to DNA. Many conventional cancer therapies target microtubular stability or DNA damage, and whilst there is evidence in the scientific literature that drugs targeting both processes might interact, the reason why has been unclear.
|
|
“Both the technique and the data it produces are likely to be a very valuable resource to the scientific community in the future,” adds Dr Carazo Salas. “It allows us to shine a light into the black box of the genome and learn exciting new information about the basic building blocks of life and the complex ways in which they interact.”
|
|
All the data from the study is being published online as an open resource for researchers to use. It will be available online at www.sysgro.org.
|

