| May 31, 2017 |
Enzymes engineered using direct evolution
|
|
(Nanowerk News) Organic molecules containing a fluorine atom are widely used in the materials, agrochemical and pharmaceutical industries. However, synthesizing the carbon-fluorine bond typically utilizes toxic metal catalysts and requires anhydrous conditions and high temperatures. Now an international team has developed a milder, more efficient enzyme-based approach to create this bond (Angewandte Chemie, "Directed evolution of a fluorinase for improved fluorination efficiency with a non-native substrate").
|
|
This enzymatic method works in aqueous conditions and at mild temperatures and was developed by a consortium that included researchers from A*STAR and other institutions. “These conditions are really highly attractive,” explains team member Yee Hwee Lim from the A*STAR Institute of Chemical and Engineering Sciences.
|
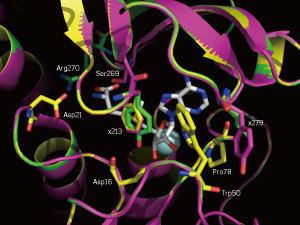 |
| A close-up of the SAM-binding sites of the native enzyme (yellow) and two mutants (magenta and green). The mutated positions 213 and 279 are labeled. (© Wiley)
|
|
The team utilized one of the highly specific fluorinase enzymes that are found in nature — FlA1. These enzymes catalyze the formation of a carbon-fluorine bond in S-adenosylmethionine (SAM) using inorganic fluoride via a substitution mechanism.
|
|
The enzyme, while excellent at catalyzing fluorination reactions with this natural molecule, did not work well on non-natural molecules. Moreover, efforts to modify the enzymes’ structures and, thus, function had been a struggle — until now.
|
|
“We showed for the first time that the fluorinase enzyme can be engineered, and that the engineering can improve its enzymatic activity even on non-natural molecules,” Lim says.
|
|
The team utilized the established technique known as ‘directed evolution’ which mimics natural selection to evolve enzymes so they can react well with non-natural molecules.
|
|
“Directed evolution has never been successfully applied to this enzyme, until now,” says Lim. “This is a difficult enzyme to work with and we faced a lot of challenges including issues with product degradation.”
|
|
The team used the radiolabeling of 5’-chloro-5’-deoxyadenosine (5’-CIDA) to showcase their capabilities. In this two-step reaction, radiolabeled 5’-CIDA is converted to SAM, and then fluorinated to form 5'-fluoro-5'-deoxyadenosine (5'-FDA). This labeled 5’-FDA product could potentially be used for a medical diagnosis procedure known as positron emission tomography (PET).
|
|
“We show that improving an enzyme’s activity three-fold can open up more applications,” says Lim. “The native enzymes have been used previously to try and label PET agents but the reaction times were long, sometimes hours. Using our enzyme we could do the reaction in 30 minutes, a more reasonable timeline given that Fluorine-18's half-life is less than two hours.”
|
|
Lim’s team is now exploring how mutating the enzyme’s structure changes its interaction with 5’-CIDA and SAM. “I’d like to engage computational biologists to better understand more about the mutations that we have made and how those correlate with our reactions.”
|

