| Sep 17, 2013 |
Team wins second $10 million award for research in molecular programming
|
|
(Nanowerk News) During the past century, programmable technologies evolved from spinning gears and vacuum tubes to transistors and microchips. Now, a group of Caltech researchers and their colleagues at the University of Washington, Harvard University, and UC San Francisco are exploring how biologically important molecules—like DNA, RNA, and proteins—could be the next generation of programmable devices.
|
|
Erik Winfree, professor of computer science, computation and neural systems, and bioengineering, and his collaborators at Caltech and the University of Washington began the Molecular Programming Project (MPP) in 2008 as an NSF Expeditions in Computing to develop practices for programming biomolecules—much like a computer code—to perform designated functions. Over the past five years, the researchers have programmed DNA to carry out a number of tasks, from solving basic math and pattern-recognition problems to more mechanical tasks like programming RNA and DNA to selectively amplify fluorescent signals for biological microscopy. Through these initial experiments, the researchers have shown that it is possible to systematically encode specialized tasks within DNA molecules.
|
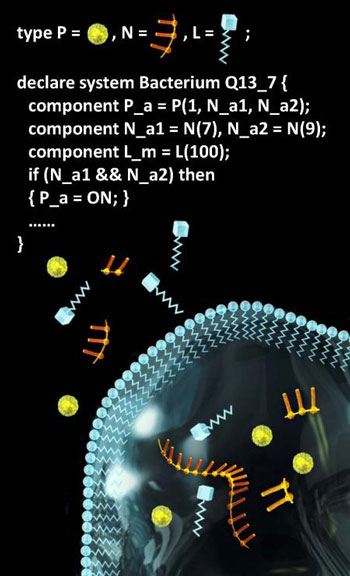 |
| Conceptual representation of programming languages for specifying molecular components and molecular systems, part of the Molecular Programming Project. (Image: Lulu Qian, Caltech)
|
|
"Computer science gave us this idea that many tasks can actually be done with different types of devices," Winfree says. For example, a 19th-century cash register and a 21st-century computer can both be used to calculate sums, though they perform the same task very differently. At first glance, writing a computer program and programming a DNA molecule may seem like very different endeavors, but "each one provides a systematic way of implementing automated behaviors, and they are both based on similar principles of information technology," Winfree says.
|
|
Expanding the team to include five additional faculty who bring expertise in structural and dynamic DNA nanotechnology, synthetic biology, computer-aided design, programming languages, and compilers, Winfree and his colleagues recently received a second Expeditions in Computing award to take their work in molecular programming to the next level: from proof-of-principle demonstrations to putting the technology in the hands of users in biology, chemistry, physics, and materials science.
|
|
The researchers aim to use molecular programming to establish general-purpose, reliable, and easy-to-use methods for engineering complex nanoscale systems from biomolecules. In the hands of users, these methods could be used to create novel self-assembling electronic and optical devices, powerful nanoscale tools for the study of biology, and programmable molecular circuits for the diagnosis and treatment of disease. In one application, the researchers hope to program DNA molecules to carry out recognition and logical circuitry for exquisitely targeted drug delivery, thus reducing drug side effects and increasing efficacy.
|
|
Today, the largest synthetic molecular programs—human-designed sequences of the A, T, C, and G bases that make up DNA—contain on the order of 60,000 bases. "That's comparable to the amount of RAM memory in my first computer, a 1983 Apple II+," says Winfree. Designed systems in the future will only become more complex, a challenge that MPP researchers aim to tackle by approaching biological systems with something computer scientists call the abstraction hierarchy.
|
|
"In some sense computer science is the art of managing complexity, because you design things that have billions of components, but a single person simply cannot understand all the details and interactions," he says. "Abstraction is a way of hiding a component's details while making it easy to incorporate into higher-order components—which, themselves, can also be abstracted. For example, you don't need to know the details of a multiplication circuit in order to use it to make a circuit for factoring." In the molecular world, the task might be different—like transporting a molecular cargo to a designated location—but abstraction is still essential for combining simpler systems into larger ones to perform tasks of greater complexity.
|
|
"Over the next several decades, the MPP seeks to develop the principles and practice for a new engineering discipline that will enable the function of molecules to be programmed with the ease and rigor that computers are today, while achieving the sophistication, complexity, and robustness evident in the programmable DNA, RNA, and protein machinery of biology," says Niles Pierce, professor of applied and computational mathematics and bioengineering at Caltech and member of the MPP.
|
|
To integrate these fields, the MPP has brought together an interdisciplinary team of computer scientists, chemists, electrical engineers, physicists, roboticists, mathematicians, and bioengineers—all of whom have a strong research interest in the intersection of information, biology, and the molecular world. The team will explore the potential of molecular programming from many perspectives.
|
|
"Because of the diverse expertise that is required to work on these challenges, the participating students and faculty come from an unusual array of fields," Pierce says. "It's a lot of fun to be in a room with this group of people to see where the discussions lead."
|
|
The 2013 Expeditions award was granted for the proposal "Molecular Programming Architectures, Abstractions, Algorithms, and Applications". Winfree and Pierce are joined on the project by four other collaborators at Caltech: Jehoshua (Shuki) Bruck, Gordon and Betty Moore Professor of Computation and Neural Systems and Electrical Engineering; Richard Murray, Thomas E. and Doris Everhart Professor of Control and Dynamical Systems and Bioengineering; Lulu Qian, assistant professor of bioengineering; and Paul Rothemund, senior research associate in bioengineering, computing and mathematical sciences, and computation and neural systems. Other collaborators include Eric Klavins and Georg Seelig from the University of Washington, Peng Yin and William Shih from Harvard, and Shawn Douglas from UC San Francisco.
|

