| Oct 29, 2013 |
Nanotechnology researchers test for drug toxicity with iPod (w/video)
|
|
(Nanowerk News) Accurate and rapid testing for drug toxicity just became easier, thanks to a half-dozen Rice University student interns working at Houston-based startup Nano3D Biosciences (n3D).
|
|
The bioengineering and nanoscale physics students just wrapped up a yearlong effort to aid the company in developing a new method for conducting high-throughput, in vitro cytotoxicity assays. A research paper about the new method was published this month in Nature’s open-access journal Scientific Reports ("A high-throughput three-dimensional cell migration assay for toxicity screening with mobile device-based macroscopic image analysis").
|
|
“This would not have been possible without the students,” said Glauco Souza, n3D’s president and chief scientific officer. “They helped develop the scientific protocols and hardware for this, and they wrote both the iPod app and the analytic software.”
|
|
Nano3D Biosciences President Glauco Souza, center, is joined by members of the team that developed a high-throughput method for in vitro cytotoxicity assays that uses a free iPod app and analytical software for automated data collection and analysis. From left are Rice University graduate students Will Haisler and Shane Neeley, senior scientist Hubert Tseng '13 and Rice graduate student Jianbo Chen.
|
|
The new assay method, which n3D has dubbed the “BiO Assay,” uses a free iPod app to collect time-lapse images of 3-D cell cultures that have been exposed to varying levels of a drug. Those images are then fed through an analytical program that measures each sample and creates time-lapse movies, graphs and charts of the drug’s cytotoxic profile.
|
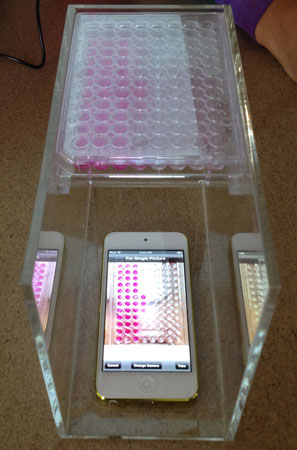 |
| A half-dozen Rice University student interns have helped Houston-based startup Nano3D Biosicences develop a high-throughput method for in vitro cytotoxicity assays that uses a free iPod app and analytical software for automated data collection and analysis.
|
|
“This literally collects about 100,000 data points during a 12-hour, overnight experiment,” said study co-author Shane Neeley, a Rice bioengineering graduate student who has interned at n3D for nine months. “That’s all relevant publishable data that relate to the different times, doses and cell types and other key variables in the experiment.”
|
|
Souza and Rice faculty members Tom Killian and Robert Raphael co-founded n3D in 2008 based on technology they created to grow 3-D cell cultures using magnetic levitation. The technology relies on inert, nontoxic magnetic nanoparticles that attach to living cells. Magnets can then be used to lift and suspend the cells as they grow and divide.
|
|
The research is part of a growing trend to create better lab techniques for testing drug toxicity. At issue is the fact that the toxic side effects of many new drugs are discovered only during human clinical testing, which means tests on 2-D cell cultures and on lab animals failed to identify the toxicity risk in humans. Cells grown in 3-D cultures behave more like the body’s native tissues, and scientists have scrambled to find ways of using 3-D cultures to reduce the need for animal testing and to rule out toxic drug candidates earlier.
|
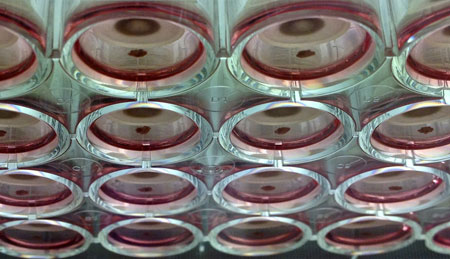 |
| N3D's technology grows 3-D cell cultures using magnetic levitation. The technology relies on inert, nontoxic magnetic nanoparticles that attach to living cells. Magnets can then be used lift and suspend the cell cultures as they grow.
|
|
“It’s been estimated that improving the accuracy of early cytotoxicity screenings by even 10 percent could save drug companies as much as $100 million per drug,” said study co-author Hubert Tseng ’13, n3D’s senior research scientist. Tseng, who interned with the company prior to earning his Ph.D. in bioengineering in March, played an instrumental role in developing several of the company’s products, including the BiO Assay.
|
|
Souza said the company developed the BiO Assay out of necessity; Interns in the lab were spending hour after hour snapping photos of individual cell cultures on the microscope. Each experiment involved exposing a hundreds of cell cultures to varying doses of a drug. The microscopic images revealed how much smaller the culture became over time, as the toxic drug slowly killed off the cells in the colony. Each culture was grown in its own tiny chamber on standard plates that each contained 96 chambers.
|
|
“Without looking in the microscope, just looking at the camera and clicking like a robot, it would take 20 minutes to take pictures of all 96 wells on one plate,” Souza said. “To analyze that, all 96, with a ruler, took even longer.”
|
|
Study first authors David Timm ’12 and Jianbo Chen ’13, professional masters students in nanoscale physics, had to repeat that tedious process over and over, as often as possible, on dozens of 96-well plates that were being used in multiple experiments.
|
|
“We decided there had to be a better way, so we began experimenting with using an iPod,” Souza said. “It was promising, but none of the available apps worked very well, so we decided we needed to make our own. I called Apple and asked them to give me the name of a developer here in Houston. When they heard where I was, they said, ‘Don’t (hire a developer). Go to Rice University and get a couple of students instead. You’ll get a better app, and it will do exactly what you want.’”
|
|
|
|
Study co-author William Haisler ’12 created the iPod app, which can snap photos every few seconds for days at a time. Neeley wrote analytical software processes the images Additional study co-authors include Killian, professor and chair of physics and astronomy; Raphael, professor of bioengineering; former Rice undergraduate David Sing ’11; Jacob Gage of n3D; and Mehdi Dehghani and Kevin Rosenblatt, both of the University of Texas Health Science Center at Houston. The research was supported by the National Science Foundation’s Small Business Innovation Research program and the Texas Emerging Technology Fund.
|


