| Posted: Aug 25, 2009 | |
Visualizing the DNA helix with cryoEM |
|
| (Nanowerk Spotlight) Recent advances have made cryo-electron microscopy (cryoEM) an important imaging tool for major applications in both medicine and nanobiological research. In this form of electron microscopy the sample is studied at cryogenic temperatures, i.e. below -150°C. Researchers can use cryoEM to visualize a broad range of assemblies or nanometer-scale structures at near-atomic resolution and in three dimensions. This imaging method covers a scale range from tens of micrometers down to single angstroms and provides valuable structural information for numerous scientific disciplines including structural biology, cell biology, medical and pharmaceutical science. Early last year, a research team used cryoEM to capture a three-dimensional image of a virus at a resolution of 4.5 angstroms ("New technique takes a big step in examination of small structures "). The technique also has the added benefit of maintaining the sample being studied in a state very similar to its natural environment. Other imaging techniques used regularly, such as X-ray crystallography, require the sample be manipulated. | |
| A Japanese-UK research team has now demonstrated that cryoEM image analysis may be exploited to obtain structural information of sufficient resolution to reveal the absolute three-dimensional (3D) configuration of a designed DNA nanostructure. With their technique they have obtained structural information at sufficient resolution to visualize the DNA helix and reveal the absolute stereochemistry of a self-assembled DNA tetrahedron. Each edge is a 7 nm, 20 base pair duplex, and the edges are connected covalently through single unpaired adenosine nucleotides, making it a rigid, triangulated structure that could serve as a building block for larger 3D structures or as a molecular cage. This DNA tetrahedron is the smallest 3D nanostructure made by DNA self-assembly. | |
 |
|
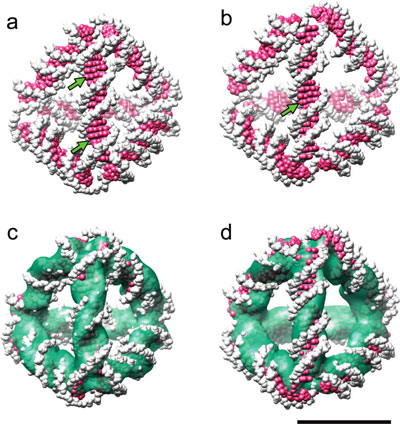
| Comparison between the 3D density map and structural models of the two expected diastereomers. (a,b) Space-filling representation of atomic models of diastereomers with (a) the minor groove and (b) the major groove facing outward at the edge center. Arrows indicate the major grooves of the double helix, which are resolved in the density map. (c,d) Superposition of the model structures (a,b) and the density map obtained by cryoEM image reconstruction. Scale bar, 5 nm. (Reprinted with permission from American Chemical Society) | |
| "We have achieved resolution sufficient not only to verify the DNA tetrahedron's structure but also to discriminate between structurally similar diastereomers, demonstrating that cryoEM is an indispensable tool for the characterization of 3D nanostructures designed to be self-assembled from biomolecular components," say the authors, led by Keiichi Namba a professor at Osaka University's Graduate School of Frontier Sciences, and Andrew J. Turberfield professor of physics at Oxford University. | |
| The researchers have reported their findings in a recent issue of Nano Letters ("High-Resolution Structural Analysis of a DNA Nanostructure by cryoEM"). | |
| Since scientists have discovered DNA as an ideal building material for bottom-up nanofabrication, various studies have already shown the complexity of architecture that is achievable using DNA as building blocks. For instance, nanofabrication via molecular self-assembly has already resulted in simple DNA polyhedra with connectivities of a cube, octahedron, tetrahedron and even icosahedron (see: "Self-assembled DNA nanocapsules for drug delivery"). A wide range of potential applications for these DNA cages have been proposed, including drug delivery, protein crystal templates, medical diagnosis, computing, and DNA nanorobotic systems (see: "The gripping potential of DNA nanotechnology "). | |
| For practical applications it is essential to verify that the 3D structures are accurately produced as designed, which requires high resolution, at least sufficient to resolve the DNA helix. | |
| "Examination and confirmation of accurate 3D DNA constructions are particularly challenging given their extremely small sizes" say Namba and Turberfield. "Structures have been visualized by either atomic force microscopy (AFM) or electron cryomicroscopy (cryoEM). AFM is a powerful technique for high-resolution observation of flat structures on flat substrates but less well adapted to the visualization of 3D structures of nonflat molecules such as polyhedra. Previously, 3D images of DNA polyhedra have been reconstructed by cryoEM and single particle image analysis to demonstrate that their overall structures are consistent with their designs, but the resolution of the reconstructions is still too low to determine absolute 3D configurations or to distinguish stereoisomers even for relatively large particles with sizes ∼20-80 nm." | |
| According to the scientists, the resolution of cryoEM 3D image reconstruction can be increased by increasing the number of particle images to be averaged – a resolution better than 4 Å has already been reached by aligning and averaging about 10 000 particle images – but the achievable resolution is still dependent upon the particle size because the low contrast and low signal-to-noise ratio of raw cryoEM images make it difficult to select particles and accurately align raw images for averaging. This difficulty becomes extremely serious for particles smaller than a critical size of around 10 nm. | |
| Naba and Turberfield explain that they "hypothesized that image noise in the solvent region surrounding the DNA tetrahedron molecule had lead to misclassification and misalignment of the particle images to some extent, resulting in the relatively poor resolution. We therefore applied a solvent flattening technique to reduce the noise in the solvent region and to improve the accuracy of image classification and alignment in the refinement cycle of single particle image analysis." | |
 |
|
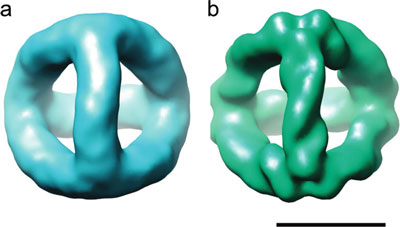
| 3D density maps of the DNA tetrahedron revealed by cryoEM image reconstruction. (a) 3D density map at 20 Å resolution produced by using a simple circular mask to isolate particle images. (b) 3D density map at 12 Å resolution produced by using a tight mask determined from the density map shown in panel a. Scale bar, 5 nm. (Reprinted with permission from American Chemical Society) | |
| It turned out that the solvent flattening technique was very effective, possibly as a result of the presence of a large, solvent-filled space within the DNA tetrahedron as well as the significant noise reduction in the solvent region surrounding the particle and the resolution of the 3D density map was dramatically improved. The resolution of the 3D density map was dramatically improved to 12 Å – the helical twist of the DNA edges can now be seen clearly, and the exact positions of the B-DNA major grooves can be identified (see above image). | |
 By
Michael
Berger
– Michael is author of three books by the Royal Society of Chemistry:
Nano-Society: Pushing the Boundaries of Technology,
Nanotechnology: The Future is Tiny, and
Nanoengineering: The Skills and Tools Making Technology Invisible
Copyright ©
Nanowerk LLC
By
Michael
Berger
– Michael is author of three books by the Royal Society of Chemistry:
Nano-Society: Pushing the Boundaries of Technology,
Nanotechnology: The Future is Tiny, and
Nanoengineering: The Skills and Tools Making Technology Invisible
Copyright ©
Nanowerk LLC
|
|
|
Become a Spotlight guest author! Join our large and growing group of guest contributors. Have you just published a scientific paper or have other exciting developments to share with the nanotechnology community? Here is how to publish on nanowerk.com. |
|
