| Posted: Nov 25, 2009 | |
Counting single molecules inside nanodroplets |
|
| (Nanowerk Spotlight) A lot of the scientific knowledge in chemistry and biology comes from experiments on ensembles of molecules by which a vast number of duplicate behaviors are investigated and averaged responses are recorded. Especially the ability to make measurements at the single molecule level provides crucial information about biological and chemical systems. This research depends on molecular manipulation technologies that are able to isolate individual molecules and sequentially transport them for measurement and, potentially, manipulation. | |
| To this end, generation and manipulation of small, highly monodisperse droplets have received a lot of attention in the biotech community recently. Using a simple droplet generator chip, scientists can generate millions of droplets – each containing a single biomolecule such as DNA or RNA – in a short amount of time. Each individual droplet is isolated from another droplet, hence, meaning that a million droplets constitute a million individual microreactors running a million reactions simultaneously. However, the true potential of the droplet platform can be realized only if one can use a sensitive detection technique to be able to 'see and count' what is present in each individual droplet at single molecule level. | |
| Researchers have now, for the first time, demonstrated direct and amplification-free single molecule detection of biomolecules in sub-nanolitre droplets through application of Cylindrical Illumination Confocal Spectroscopy (CICS) and droplet confinement within a retractable microfluidic constriction. | |
| Typically, researchers working with droplet microfluidics use CCD cameras for fluorescence detection. These cameras lack the sensitivity required to detect low concentration of fluorescent molecules present within droplets. As a result for conducting any target specific assay some type of amplification scheme like a PCR assay is required. | |
| "In our work, we address this issue through innovative use of microfluidics and a modified form on confocal spectroscopy," Jeff Tza-Huei Wang, an associate professor in mechanical & biomedical engineering at Johns Hopkins University, tells Nanowerk. "As demonstrated in our work, the design we proposed can achieve close to 100% molecular detection efficiency i.e. count almost all the fluorescent molecules present in every single droplet. This means ultimate form of droplet differentiation, as we can differentiate between small amount of molecular content difference over droplets, which is not possible with other detection techniques." | |
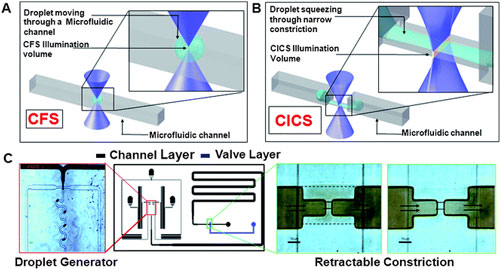 |
|
| A droplet moving through a microfluidic channel is shown. Inset shows the size of the droplet relative to the illumination volume of a standard Confocal Fluorescence Spectroscopy (CFS) setup. B. In contrast, elongation of a droplet squeezing through a microfluidic constriction is shown. The inset shows the sheet-like illumination volume of a Cylindrical Illumination Confocal Spectroscopy setup relative to the elongated droplet in the microfluidic constriction. C. The multilayered microfluidic device designed for the experiments in this paper is shown. A flow focusing geometry was used for droplet generation. The left panel shows droplets being generated using a food dye as the discrete phase. The rightmost panels show the retractable constriction region in either the open (left) or actuated (right) state, as described in text. (scale bar: 50 µm). (Reprinted with permission from The Royal Society of Chemistry) | |
| Wang and his team report their findings in a recent issue of Lab on a Chip ("Counting single molecules in sub-nanolitre droplets" – free access article). The beauty of this technique lies in its simplicity, which allows them to perform quantification within droplets, without the requirement of any time-consuming and complicated amplification techniques. This technique can have immediate application in single cell studies, where the differences between individual droplets are expected to be very small. | |
| The researchers use a retractable valve that constricts part of a microfluidic channel. This stretches the droplet without breaking it and slows its passage through the narrowed channel. The altered droplet shape and extra time allows the trapped biomolecules inside to be detected using fluorescence spectroscopy. Using the technique, the team were able to detect nucleic acids inside droplets. | |
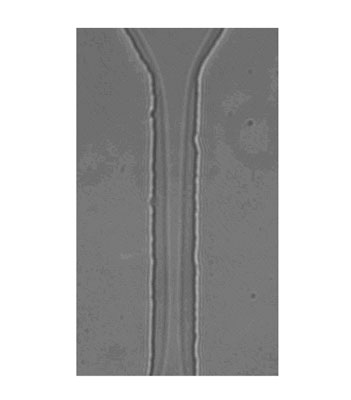 |
|
| A high magnification image of a droplet squeezing through a 20 µm by 10 µm constriction. This lateral view clearly shows the continued presence of the continuous phase surrounding the droplet as it is pushed through the constriction. (Reprinted with permission from The Royal Society of Chemistry) | |
| With their novel technique, Wang's team overcomes challenges involved with biomolecular quantification in droplets, including short intradroplet signal acquisition times and droplet?illumination volume size mismatches. | |
| "In our platform, one-dimensional beam shaping using cylindrical optics CICS produces a sheet-like illumination volume, maximizing intra-droplet detection efficiency," explains Wang. "Furthermore, droplet confinement through a microfluidic constriction extends droplet duration through the illumination volume, providing the spatial and temporal resolution necessary to detect single biomolecules." | |
| He gives an example where the new technique could assists researchers in conducting amplification-free, high-throughput single cell studies: A clonal population of cells can be genetically identical across the population, but still exhibits significant variation in phenotypic characteristics at single cell level. As a result, there is a strong interest in the biological community to understand the origins of this variability at single cell level. Droplet microfluidics has recently been applied towards such high throughput studies at single cell level. However, these studies are limited to amplification schemes like PCR due to lack of a sensitive detection platform. Wang is confident that their new platform will be able to break this barrier and make it possible to conduct amplification free single cell studies within droplets. | |
| The researchers at Johns Hopkins already demonstrated the application of the single molecule counting technique within droplets with a sample biochemical system of stained DNA molecules. | |
| "However" says Wang, "the system must be optimized with due consideration to throughput and sensitivity required for the particular application being studied. So we envision the future research in this area to be geared towards rigorous characterization of the applicability of this technique towards a variety of biochemical applications." | |
 By
Michael
Berger
– Michael is author of three books by the Royal Society of Chemistry:
Nano-Society: Pushing the Boundaries of Technology,
Nanotechnology: The Future is Tiny, and
Nanoengineering: The Skills and Tools Making Technology Invisible
Copyright ©
Nanowerk LLC
By
Michael
Berger
– Michael is author of three books by the Royal Society of Chemistry:
Nano-Society: Pushing the Boundaries of Technology,
Nanotechnology: The Future is Tiny, and
Nanoengineering: The Skills and Tools Making Technology Invisible
Copyright ©
Nanowerk LLC
|
|
|
Become a Spotlight guest author! Join our large and growing group of guest contributors. Have you just published a scientific paper or have other exciting developments to share with the nanotechnology community? Here is how to publish on nanowerk.com. |
|
