| Posted: Mar 28, 2011 | |
Rapid probing of single molecules with a nanomechanical interface |
|
| (Nanowerk Spotlight) The complex processes inside living systems emerge from the interactions of countless molecules. Traditionally, a lot of the scientific knowledge in chemistry and biology comes from experiments on ensembles of molecules by which a vast number of duplicate behaviors are investigated and averaged responses are recorded. However, understanding these interactions at the single molecule level is of great importance because mechanisms governing their function can be revealed best by interrogating individual molecules. | |
| Experiments with large number of molecules tend to be easier, however important details get lost in the crowd. For this reason, scientists rely on single-molecule techniques that allow them to isolate individual molecules and sequentially transport them for measurement and, potentially, manipulation. | |
| Most commonly, molecules are interfaced with tools like optical tweezers and atomic force microscopes. These devices are precision force sensors. When a molecule goes through a structural change – for example, when it binds to another molecule – the forces generated as a result of this change can be detected. In some cases, the same tools can be used to apply force to a molecule to see when it stops functioning. These measurements allow scientist to "reverse engineer" biological molecules. | |
| Many interactions among biological molecules are short lived. Their lifetimes can be as short as a billionth of a second (a nanosecond). The existing single molecule techniques are limited in their temporal resolution, probing the timescales on the order of several milliseconds to a second. | |
| In new work, researchers at The Rowland Institute at Harvard and Harvard University have extended the reach of single molecule experiments to the microsecond timescale. | |
| "In general, this allows probing events that last only a few microseconds" Ozgur Sahin explains to Nanowerk. "To put it simply, if an interaction of interest lasts a few microseconds, you have a very short window of time to probe it. If the measuring device is slow, the interaction disappears before the measurement takes place. We have developed a technique to probe these interactions with extremely short durations." | |
 |
|
| False color electron microscopy image of a T-shaped cantilever designed for single-molecule experiments. (Image: Ozgur Sahin, Harvard University) | |
| Sahin's approach is based on his group's previous work on atomic force microscope technology (see our previous Nanowerk Spotlight: "Feeling your way through the nanoworld "). The team has now developed T-shaped cantilevers that can measure intermolecular forces rapidly, increasing the rate of single-molecule force spectroscopy by nearly four orders of magnitude. | |
| The new design is using the torsional harmonic cantilever concept that was developed for high-resolution nanomechanical analysis of samples (described in above Spotlight). These T-shaped cantilevers generate high-speed force-distance curves during the tapping-mode AFM imaging process. | |
| "The twisting oscillations of this type of cantilevers can respond to quick changes in molecular forces" says Sahin. "We took advantage of this and used the T-shaped cantilever to probe rapid chemical interactions between molecules." | |
| Sahin and first author Mingdong Dong report their findings in the March 22, 2011 edition of Nature Communications ("A nanomechanical interface to rapid single-molecule interactions"). | |
 |
|
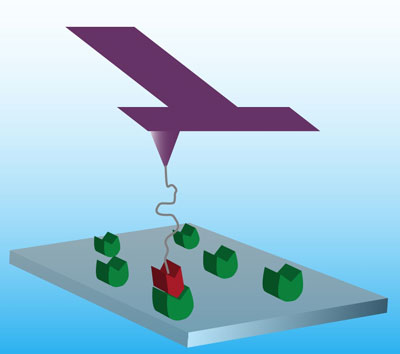
| Schematic of single-molecule force spectroscopy on the microsecond timescale. The ligand is attached to a specially designed T-shaped AFM cantilever via a flexible polymer spacer. Tip-sample interactions twist the vibrating cantilever. The vertical deflection signal gives the tip position, and the twist angle gives the instantaneous force on the tip. (Image: Ozgur Sahin, Harvard University) | |
| The response of single molecules to external forces depends on the duration of the external force. If pulled gently, chemical bonds between molecules last long before they break apart. Molecules generate larger forces if they are pulled apart faster. | |
| Sahin notes that in their experiments, they have seen a dramatic increase in the forces supplied by molecules. | |
| "In this process, the rate of increase in the forces carries information about the energy barriers holding the molecules together" he says. "Our data shows clear evidence for an additional energy barrier in the interaction of biotin and streptavidin molecules. Computer simulations previously made by another group have indeed suggested the existence of an additional barrier, but there wasn't any conclusive experimental evidence." | |
| An indirect benefit of rapid measurements is that it naturally produces images based on molecular measurements. Because the forces bear specific information about the molecules, these images can be used to locate certain molecules of interest on a surface with precision. | |
| Examples of entire force waveforms and force-distance curves. Tip-sample interactions recorded during the experiments are plotted against time and tip-sample distance. The curves are plotted 300 times slower than the actual recording rate. A pause equal to 300 ms is added right after curves that exhibit pulling forces more than 200 pN. | |
| The immediate application of this technique is in biological imaging. There are many questions related to the structure and organization of cellular membranes where locations of membrane proteins relative to each other and relative to underlying cellular structures are unknown. The specificity of intermolecular forces allows one to locate certain types of molecules on a surface, which may well be the cell membrane. Therefore, imaging cellular membranes to identify patterns in the organization of membrane receptors is one area where important advances can be expected. | |
| In addition to imaging applications, deformation and failure mechanisms of structural proteins can be investigated. These processes depend on the timescale of external forces. "A molecule can lose its structural integrity in a different way at high speeds – such as during shock or impact – than at low speeds. Understanding these mechanisms is important for the design of biomimetic materials with improved performance," says Sahin. | |
| He points out that there have been major advances in nanoscale experimental methods in the past decade. "While mechanically and electronically demanding aspects of these methods are currently being addressed, the ability to manipulate molecules under investigation in a simple and robust way is limited. We and others rely on biochemical methods that are not particularly suitable for nanoscience. Consequently, trial and error have been important for experimental nanoscience." | |
 By
Michael
Berger
– Michael is author of three books by the Royal Society of Chemistry:
Nano-Society: Pushing the Boundaries of Technology,
Nanotechnology: The Future is Tiny, and
Nanoengineering: The Skills and Tools Making Technology Invisible
Copyright ©
Nanowerk LLC
By
Michael
Berger
– Michael is author of three books by the Royal Society of Chemistry:
Nano-Society: Pushing the Boundaries of Technology,
Nanotechnology: The Future is Tiny, and
Nanoengineering: The Skills and Tools Making Technology Invisible
Copyright ©
Nanowerk LLC
|
|
|
Become a Spotlight guest author! Join our large and growing group of guest contributors. Have you just published a scientific paper or have other exciting developments to share with the nanotechnology community? Here is how to publish on nanowerk.com. |
|
