| Posted: Jun 06, 2011 | |
Novel approach for highly sensitive detection of miRNA down to individual strands |
|
| (Nanowerk Spotlight) MicroRNAs (miRNAs) are short ribonucleic acid (RNA) molecules, consisting of 21-25 nucleotide bases, that negatively regulate gene expression, also termed as gene silencing. Each miRNA is thought to regulate multiple genes, and since the human genome encodes hundreds of miRNAs, the potential regulatory circuitry afforded by miRNA is enormous. | |
| Although the first published description of a miRNA appeared ten years ago, only in the last two to three years has the breadth and diversity of this class of small, regulatory RNAs been appreciated. A great deal of effort has gone into understanding how, when, and where miRNAs are produced and function in cells, tissues, and organisms. | |
| Recent discoveries suggest the association of specific miRNA sequences with a spectrum of diseases including cardiovascular and autoimmune diseases, as well as with a variety of cancers. It is therefore imperative, for diagnostics and prognostics, to accurately measure the expression levels of target miRNA molecules in patients' tissue samples or body fluids. | |
| Commercial microarrays for miRNAs use either fluorophores or immunofluorescent tags such as streptavidin-phycoerythrin as labeling agents, and involve multiple-step protocols. In addition to their lengthy procedures, the suggested hybridization time for the commercial microarrays takes approximately 16–18 hours, which is unrealistically long, especially for point-of-care applications. | |
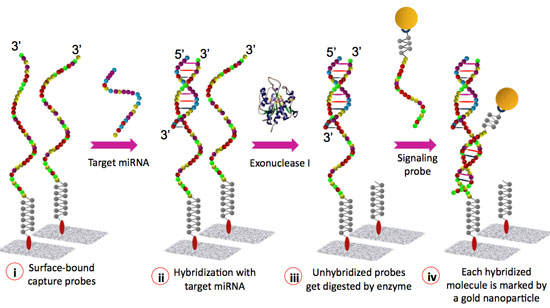 |
|
| Figure 1: Schematic representation of the miRNA detection assay. (Image: Institute of Bioengineering and Nanotechnology) | |
| "To circumvent the limitations associated with fluorescent labeling and image scanning, it was imperative to exploit alternative ways for the direct analysis of miRNAs in an array format," Somenath Roy, a research scientist at the Institute of Bioengineering and Nanotechnology (IBN) in Singapore, tells Nanowerk. "Apart from reducing the technical complexities, we also aimed to reduce the turnaround time and the cost of the assay." | |
| Roy is first author of a paper in a recent edition of Lab on a Chip ("A microfluidic-assisted microarray for ultrasensitive detection of miRNA under an optical microscope") where he and IBN colleague Jun Hui Soh, together with Zhiqiang Gao from the National University of Singapore, demonstrate fast and ultrasensitive detection of specific miRNAs. | |
| Although conventional microarray analyses can monitor global changes in gene expression, these studies are limited by lengthy and complex assays. IBN's microarray is sensitive enough to detect low copy numbers in an unamplified and unmodified total RNA sample, thereby reducing the cost, complexity and bias associated with miRNA labeling or/and PCR. | |
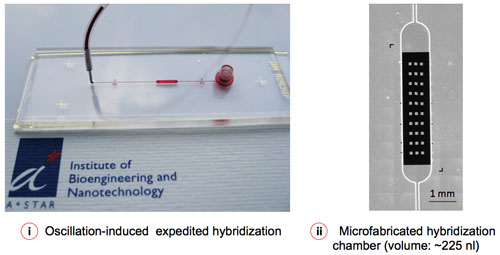 |
|
| Figure 2: Microfluidic-assisted sample delivery and oscillation-hybridization reduces the reagent cost and assay time to a significant extent. (Image: Institute of Bioengineering and Nanotechnology) | |
| Roy explains that the existing methods of quantifying miRNAs include Northern Blot, reverse-transcription polymerase chain reaction (RT-PCR) and more recently, fluorescent microarrays. While Northern Blot is more conventional and a gold standard for miRNA quantification and RT-PCR is the most sensitive method thus far, both of these techniques suffer from specific limitations, in addition to their common problem of extensive sample preparation. | |
| While a slight degradation – even a single cleavage – in the RNA sample severely affects quantitative expression for Northern Blot analysis, RT-PCR is extremely prone to contamination due to its inherently high sensitivity. The state-of-the-art fluorescent microarrays are unrivalled in terms of parallelism and throughput, yet are not suitable for point-of-care applications, primarily due to their requirement for target amplification, labeling and long hybridization time (usually 16-18 hours). | |
| Roy and his collaborators now have designed and implemented a microfluidic-assisted microarray, which addresses some of the aforesaid limitations by reducing technical complexities and turnaround time as well as the cost of the assay. | |
| "First, with our novel microarray, there is no need for target amplification or labeling for quantifying specific miRNAs from total RNA samples, thereby reducing the assay time and complexity significantly" says Roy. "Second, our platform requires only 1-5 ng of total RNA in 5–10 µl of sample, compared to 1-10 µg of starting total RNA or the equivalent of enriched miRNA required by conventional microarrays. Third, the microfluidic-assisted sample delivery and flow convection reduces the assay time from 16-18 hours to a mere 1 hour, which is especially important for point-of-care applications. Finally, the lower limit of detection (LOD) for our microarray is approximately 1 zmol (equivalent to ∼300 copies/µl), which is at least 100 times more sensitive than commercially available fluorescent microarrays." | |
| The use of sandwich assays for DNA/RNA detection has already been demonstrated (see for instance: "Gold nanoparticle-based detection of genomic DNA targets on microarrays using a novel optical detection system"). Here, after hybridizing with capture probes, the target nucleic acid molecules undergo a second hybridization with fluorescence- or nanoparticle-tagged detection probes. | |
| Roy notes that this approach, however, may lead to difficulties in reliably quantifying miRNAs due to the use of extremely short capture probes and detection probes. The specificity of the approach may also be discounted, as the risk of cross-hybridization is greatly increased owing to the inability of the extremely short probes to efficiently bind on their target miRNAs. Further, in the sandwich configuration, the absence of a spacer between the two end-to-end duplexes reduces the reliability of the microarray. | |
| In stark contrast to the conventional sandwich assay, IBN's system has been designed in such a way that the capture probe hybridizes with the entire length of its target miRNA, as well as with that of the signaling probe, thereby significantly enhancing the integrity of the assay. | |
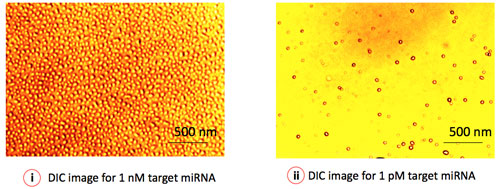 |
|
| Figure 3: Differential Interference Contrast (DIC) microscopy depicts the virtual 3D image of individual gold nanoparticles (GNP), representing a single miRNA target. The surface coverage (number of particles/area) of GNP represents the concentration of specific miRNA in the sample. (Image: Institute of Bioengineering and Nanotechnology) | |
| "In addition, our detection technique is vastly different from other nanoparticle-based microarrays that employ silver enhancement to realize nucleic acid detection" Roy points out. "Besides introducing additional process steps, silver enhancement may yield misleading results through non-specific background staining. Our direct visualization/quantification process using differential interference contrast (DIC) microscopy creates a straightforward virtual 3D image of the individual nanoparticles, conjugated to the signaling probes. Adoption of DIC imaging as the microarray detection technique offers the possibility of single molecule analysis under an optical microscope." | |
| The microarray platform developed by the scientists in Singapore may be used in point-of-care applications, where the relative expressions of a set of specific nucleic acids need to be analyzed in a timely and simplistic manner, for example, for the diagnosis and prognosis of cancer or cardiovascular disease in a doctor's clinic. | |
| Recent studies have indicated that miRNAs circulate in the bloodstream, and the relative abundance of specific miRNAs in plasma or serum can serve as biomarkers of cancer and other diseases, such as cardiovascular and autoimmune diseases. | |
| "Our ultrasensitive microarray platform may be suitable for the direct profiling of miRNAs from patient samples such as whole blood or serum," says Roy. "Efforts are under way to quantify circulating miRNAs in body fluids. Such a tool would be of great scientific value and may open a new paradigm in routine miRNA expression profiling and molecular diagnostics." | |
| He cautions, though, that due to the relatively tiny amount of circulating miRNAs and the large amount of proteins, the detection of miRNA directly from serum samples is technically challenging. Furthermore, the measurement of circulating miRNAs as biomarkers is associated with specific challenges, including those related to pre-analytical variation and data normalization, and there is a pressing need for better standardization methods. | |
 By
Michael
Berger
– Michael is author of three books by the Royal Society of Chemistry:
Nano-Society: Pushing the Boundaries of Technology,
Nanotechnology: The Future is Tiny, and
Nanoengineering: The Skills and Tools Making Technology Invisible
Copyright ©
Nanowerk LLC
By
Michael
Berger
– Michael is author of three books by the Royal Society of Chemistry:
Nano-Society: Pushing the Boundaries of Technology,
Nanotechnology: The Future is Tiny, and
Nanoengineering: The Skills and Tools Making Technology Invisible
Copyright ©
Nanowerk LLC
|
|
|
Become a Spotlight guest author! Join our large and growing group of guest contributors. Have you just published a scientific paper or have other exciting developments to share with the nanotechnology community? Here is how to publish on nanowerk.com. |
|
