| Posted: Oct 10, 2007 | |
Unlocking nature's protein material concepts enables tomorrow's nanotechnology materials and cures |
|
| (Nanowerk Spotlight) Nature is truly a brilliant nano engineer and has been so for billions of years. There is an abundance of ‘smart’ biological materials with hierarchical nanostructures - built from proteins - that are capable of adapting to new tasks, are self-healing, and can self-assemble autonomously simply out of a solution of building blocks. The performance and capability of these natural materials is something engineers can only dream of today. But by unlocking nature's secrets tiny step by tiny step, one day we will be able to not only duplicate but surpass the performance of natural materials. Only in recent years have scientists begun to understand the underlying principles and mechanisms of these materials - Why is spider silk stronger than steel? Why can cells be stretched reversibly several times of their original length? What kinds of molecular flaws lead to malfunctions in cells and tissues, as it occurs in Alzheimer’s, rapid ageing disease progeria or muscle dystrophies, diseases in which the cell or tissue fails mechanically? | |
| Scientists at MIT have, for the first time, revealed the fundamental fracture and deformation mechanisms of biological protein materials, clarifying some long-standing issues about the deformation behavior of cells and Alzheimer’s pathogens. The researchers report that the fracture mechanisms of two abundant nanoscopic building blocks of many proteins and protein materials exhibit two distinct fracture modes, depending on the speed of deformation. This is a surprising observation with far-reaching implications for the development of novel self-assembled protein materials and possibly the cure of certain genetic diseases. | |
| Paving the way for tomorrow's supermaterials, scientists need to unlock nature's structural design rules, in particular for nanoscopic hierarchical molecular structures, and make them available to engineers. This is only possibly through a deep understanding of the structure-property relations in biological materials. There is also a surprising relationship between these material design issues and the understanding (or rather lack thereof) of genetic diseases, where structural changes are due to mutations on the molecular level which lead to changed chemical and mechanical properties, which in turn lead to a malfunction of the protein network under mechanical load. | |
| "We have made a surprising discovery" Prof. Markus J. Buehler explains to Nanowerk: "Based on several different protein domains studied, we discovered that they all behave similarly. Both alpha helix (AH) and beta sheet (BS) domains (the building blocks of many proteins) exhibit two distinct fracture modes, depending on the speed of deformation." | |
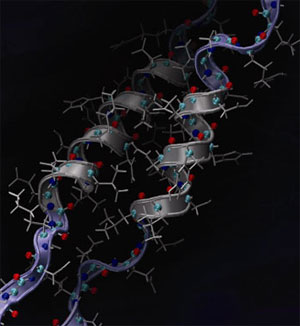 |
|
| Rupture of a coiled-coil double helical protein structure as found in the cell’s cytoskeleton, hair and hoof, under mechanical load. The snapshot shows an intermediate stage in which part of the protein’s structure is still intact (helical region), while part of the structure has been destroyed (straightened broad region). (Image: Prof. Buehler/MIT) | |
| This finding significant in order to identify the role of these proteins domains in cells and in extracellular tissue. "Biological systems can be considered as complex mechanical systems, in which displacements and forces trigger integrated chemical and biological responses," says Buehler. "In these mechanical networks, the lack of quantification of the force levels that lead to rupture of proteins or bonds between proteins has thus far precluded us from rigorously understanding the details of how these systems work." | |
| Whether or not different fracture modes of protein structures exist has been a major issue in the community. So far there has been a large gap between simulation and experiment – most probably because these two approaches have been associated with different time scales (previous simulations over several nanoseconds and experiments over seconds of time scales). As a consequence, experiment and simulation suggested different fracture mechanisms and predicted vastly different force levels that lead to rupture of the protein structures. Since experimental investigations can not provide direct evidence for the actual fracture mechanism (time scale too short, length scale too small), there has been no rigorous understanding about these fundamental processes. | |
| Buehler, the Esther and Harold E. Edgerton Professor at MIT's Department of Civil and Environmental Engineering, and his group focus on the systematic understanding of the macroscopic mechanical response of complex materials based on their fundamental, atomistic ultrastructure using large-scale, massively parallelized novel modeling techniques that provide an atom-by-atom view of the rupture events in protein materials. Back in June we already reported on some of his previous work ("Nanostructure will become design variable for materials engineers"). | |
| While the basic structure of fibrous alpha-helical proteins that form keratin, lamin and vimentin has been known since the classic works by Linus Pauling of Caltech in the 1950s, it has not been well understood how they deform and fracture, i.e. how they respond to mechanical load at an atomic level. Such knowledge could help materials scientists design improved fibers and other biomimetic materials. Even more exciting, medical researchers might use this knowledge and be able to develop new treatment options for genetic diseases. | |
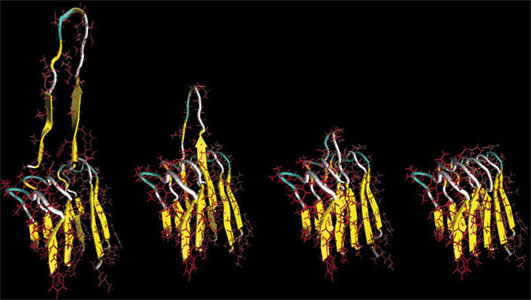 |
|
| Sequence of rupture mechanisms of an Alzheimer's amyloid protein. (Image: Prof. Buehler/MIT) | |
| With their new paper, just published in the PNAS ("Hierarchies, multiple energy barriers, and robustness govern the fracture mechanics of α-helical and β-sheet protein domains") Buehler's group has closed the gap between simulation and experiment: For the first time, thanks to large computational facilities, researchers were able to simulate the deformation mechanisms in AH and BS proteins at time scales comparable to those in regular biological function. This has been possible by carrying out simulations over extremely long time spans, approaching one microsecond. | |
| The new studies in Buehler's group have paved the way for the development of a new theoretical concept that, for the first, time allows predicting the strength of protein structures solely based on their structure and chemical bonds. The researchers used a new approach in treating the protein materials as ‘systems’, and combined this systems perspective with very large-scale full atomistic simulations. The new theory may play a similar role as the fracture theories for metals, ceramics etc. used today. | |
| "We found that structures in biological materials – compared with traditional engineering materials like metals or ceramics – are governed through a combination of strength and robustness at minimal use of building materials" says Buehler. "We have shown that the key for this behavior are hierarchies, through which soft hydrogen bonds (these are the bonds that hold water molecules together) on the nanoscale are systematically reinforced up to the macroscale. Rather than hypothesizing about these mechanisms, we have directly simulated these processes by large-scale atomistic simulation." | |
| A key finding in this research has been the discovery of two discrete fracture mechanisms that appear in protein domains. "Surprisingly we have found this in a variety of different protein structures, suggesting that this may be a universal feature" says Xuefeng Chen, an undergraduate student in Buehler's group and member of MIT’s Undergraduate Research Opportunity Program (UROP), who carried out part of the research. "In particular, we have studied AH protein domains that are part of the cell's cytoskeleton (which are responsible for the mechanical properties of cells and the nucleus, hair, wool and hoofs), as well as BS domains that appear in spider silk or amyloid fibers (appearing in Alzheimer disease)." | |
 |
|
| Molecular structure of Alzheimer’s amyloid protein. This protein structure forms crystal-like arrangements that are mechanically extremely sturdy. (Image: Prof. Buehler/MIT) | |
| The new MIT study demonstrates how it is possible to make strong materials out of extremely weak hydrogen bonds. To put this into context, hydrogen bonds are 100-1,000 times weaker than the bonds we find in steel. In fact, they are so weak that they can break simply under thermal motion (the kinetic energy of atoms and molecules is large enough so that their inertia ruptures these bonds). Buehler's study now explains how, by arranging these weak bonds in a particular hierarchical order, one can multiply their strength and obtain an effective strength that is the same as for instance in the bonds inherent in steel. In their paper the scientists have demonstrated this for the Alzheimer fibril. The key to improving their mechanical resistance is to arrange them in such a way that multiple hydrogen bonds break simultaneously under load, as it has been shown to be the case for the AH protein. | |
| "Our work also sheds light on the evolutionary driving forces that may have led to the particular geometries of the most abundant building blocks" says Buehler. "For instance, the analysis of the AH motif has shown that the structure represents a compromise between robustness and strength." | |
| "We have discovered that the particular structure of AHs may be a consequence of an adaption to generate a highly elastic structure with maximum strength," says Theodor Ackbarow, a graduate student in Buehler's group and first author of the paper. "This may explain why this protein motif is such an abundant, universally found protein motif (30% of all proteins contain the alpha helix motif)." | |
| The authors discovered that the alpha helix is 'Pareto robust,' which means that the structure is optimized only to 80% of its theoretically achievable strength. The Pareto principle is a phenomenon that is also found in many natural and economic phenomena. In this paper it is first discovered at the molecular scale. The further analysis of this surprising finding could motivate many future studies. | |
| Buehler's team notes that a wider ranging aspect of their work is the insight into what kind of structures must be used to generate high strength yet robust materials at the macroscale. From their research it becomes clear that the implementation of hierarchies is the key to achieving this goal. As scientists begin to understand these effects on the nanoscale, the impact on design and even improvement of biomimetic materials could be substantial. | |
| Understanding the structure-property link of metals and ceramics has led to the design of new high-performance materials. In the case of protein materials, similar breakthroughs could result from the theoretical advances in understanding the structure-property link for protein structures made by Buehler's group. | |
| The researchers hope that their theories, combined with new manufacturing techniques such as recombinant DNA and peptide self-assembly, will lead to the design of novel protein based materials. They also believe that their theory, due to its generic approach, could be applicable to the design of non-organic or combinations of non-organic and organic nanoscale materials such as hierarchically organized carbon nanotube bundles, nanowires or polymer-protein nanocomposites. | |
| "We hope our research will inspire scientists from a variety of disciplines (engineering, physics, chemistry, biology and medicine) and different approaches (experiment, theory and computational modeling) to work synergistically together, as only with this multi-disciplinary approach the current challenges posed by working on the nanoscale can be mastered" says Buehler. "In the future we will be able to gain a better understanding of nature's structural design rules and nature's manufacturing processes. Both will help to develop manufacturing processes which will allow the production of biological, computationally developed materials with specific properties." | |
 By
Michael
Berger
– Michael is author of three books by the Royal Society of Chemistry:
Nano-Society: Pushing the Boundaries of Technology,
Nanotechnology: The Future is Tiny, and
Nanoengineering: The Skills and Tools Making Technology Invisible
Copyright ©
Nanowerk LLC
By
Michael
Berger
– Michael is author of three books by the Royal Society of Chemistry:
Nano-Society: Pushing the Boundaries of Technology,
Nanotechnology: The Future is Tiny, and
Nanoengineering: The Skills and Tools Making Technology Invisible
Copyright ©
Nanowerk LLC
|
|
|
Become a Spotlight guest author! Join our large and growing group of guest contributors. Have you just published a scientific paper or have other exciting developments to share with the nanotechnology community? Here is how to publish on nanowerk.com. |
|
