| Posted: Oct 21, 2013 | |
Nanopore sensor could advance epigenetic cancer biomarker detection |
|
| (Nanowerk Spotlight) Epigenetic mechanisms are chemical changes in DNA that do not alter the actual genetic code but can influence the expression of genes, and can be passed on when cells reproduce. One of the most important is DNA methylation, where methyl groups – small structures of carbon and hydrogen – are appended to specific locations on a DNA strand. This plays a critical role in determining which genes are active in a cell at any given time; which in turn plays an important role in embryonic development, cell growth and reproduction, and many diseases. | |
| Numerous methods have been developed for the examination of DNA methylation in the hope of detecting biomarkers as early warning signs in various diseases, including cancer (see for instance: "Optical detection of epigenetic marks"). However, current methods for methylation analysis are costly, complex and less quantitative in determining methylation states at individual CpG sites (most DNA methylation occurs in CpG dinucleotides). | |
| Recently, both biological and synthetic nanopores have been proposed for DNA methylation detection (see for instance: "New nanopore sensor simplifies analysis of methylated DNA"). | |
| In a new report, published in the October 18, 2013, online edition of Scientific Reports ("Designing DNA interstrand lock for locus-specific methylation detection in a nanopore"), researchers employed protein nanopores to investigate a novel metal ion-bridged DNA interstrand lock, and explore its potential in location-specific methylation detection. | |
| This work provides a powerful biophysical tool to explore metal ion-nucleic acids interactions in humans and other living organisms. | |
| "The nanopore single-molecule approach presented in our work is label-free, does not require DNA sequencing, and provides a rapid, low cost and accurate tool with multiplex detection capability for epigenetic biomarker discovery and diagnostic set," Li-Qun Gu, an Associate Professor in the Department of Bioengineering and Investigator in the Dalton Cardiovascular Research Center at the University of Missouri, tells Nanowerk. "The focus of our research is single-molecule and single-base investigation of a base-pair specific metal ion/nucleic acid interaction. Such an interaction can be used to discriminate methylated and unmethylated cytosine, with a potential in DNA methylation analysis." | |
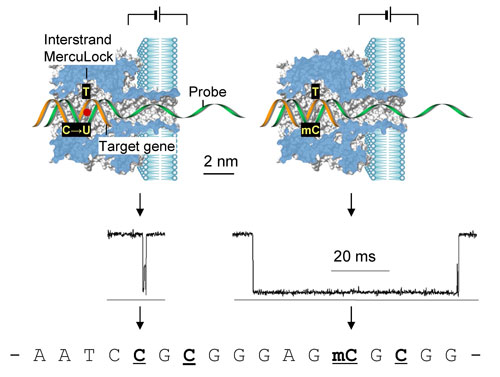
| What the team, led by Gu, found was that the uracil-thymine mismatch at a CpG site can be bound with a divalent mercuric ion. Gu explains that the metal binding creates a reversible interstrand lock, called MercuLock, which enhances the hybridization strength by nearly two orders of magnitude. | |
| "In contrast, the 5-methyl cytosine-thymine mismatch cannot form such MercuLock," he says. "Thus uracil and methylated cytosine can be discriminated from MercuLock signatures in the nanopore. Because uracil is converted from unmethylated cytosine by the bisulfite treatment, the identity of uracil corresponds to an unmethylated cytosine. Therefore the methylation status in the original gene is determined." | |
 |
|
| This scientific finding and its potential applications might be useful for the discovery of epigenetic biomarkers and cancer diagnosis. | |
| Any analytical tool that is sensitive to the change in hybridization stability can be used to detect the interstrand lock. For example, according to Professor Gerald A. Meininger, Director of the Dalton Cardiovascular Research Center at the University of Missouri, an atomic force microscope (AFM) could be used to study the dehybridization of DNA carrying multiple interstrand locks. AFM is unique in measuring the distance between interstrand locks. The sensitivity of an AFM is unique in its capability of measuring distance and force and could be capable of detecting interstrand locks, thus determining the position of each methylated and unmethylated cytosine. | |
| In this present work, though, a nanopore is utilized to study how the binding of metal ion to a single base-pair stabilizes the nucleic acids hybridization. According to Gu, this leads to three significant findings: | |
|
|
|
| "We first uncovered the binding of Hg2+ ion to the U-T mismatch, which led to the unprecedented application of mismatch-specific interstrand locks in the epigenetic detection," notes Gu. "The demonstrated ability to discriminate a single mismatched base pair bound with the metal ion renders the nanopore a powerful analytical tool to investigate the mechanism governing the metal-nucleic acids interactions." | |
| In past years, various metal-nucleic acids interactions have been extensively studied, but their application has been limited to the metal ion biosensor. A well-known example is the Mercury sensor that measures Hg2+ binding to the T-T mismatch. This work now opens an avenue to the application of metal-nucleic acids interactions in genetic and epigenetic detection. | |
| Prior to this work, researchers reported a different nanopore method for methylation detection ("Identification of epigenetic DNA modifications with a protein nanopore"). In that report, the targeted DNA was biotinylated and attached to a streptavidin for immobilization in the pore. The methylation site needs to have a fixed distance to the biotin terminal, such that upon immobilization, the methylation site is located within the sensing zone to change the nanopore current. | |
| The core of this new method developed by Gu's group is the base-specific interstrand lock. The methylation status can be determined by examining whether an interstrand lock is formed. | |
| "The 'locked' dsDNA shows greatly enhanced hybridization stability, resulting in a nanopore signature that is ~30-fold prolonged compared with dsDNA without the interstrand lock," says Gu. "Such signatures are easy to discriminate, allowing one to easily recognize the methylation status. By using a series of probes, each probe targeting a specific CpG cytosine, the methylation status for multiple CpG sites can be analyzed." | |
| The team points out that the interstrand locks they propose could find useful applications in nucleic acids nanotechnology. They can tune the hybridization strength when constructing nucleic acids nanostructures such as origami. They may also be used to improve the binding of regulators such as microRNAs to the target gene to control gene expression. | |
 By
Michael
Berger
– Michael is author of three books by the Royal Society of Chemistry:
Nano-Society: Pushing the Boundaries of Technology,
Nanotechnology: The Future is Tiny, and
Nanoengineering: The Skills and Tools Making Technology Invisible
Copyright ©
Nanowerk LLC
By
Michael
Berger
– Michael is author of three books by the Royal Society of Chemistry:
Nano-Society: Pushing the Boundaries of Technology,
Nanotechnology: The Future is Tiny, and
Nanoengineering: The Skills and Tools Making Technology Invisible
Copyright ©
Nanowerk LLC
|
|
|
Become a Spotlight guest author! Join our large and growing group of guest contributors. Have you just published a scientific paper or have other exciting developments to share with the nanotechnology community? Here is how to publish on nanowerk.com. |
|
