| Posted: Mar 08, 2016 |
New nanoparticle technology to decipher structure and function of membrane proteins
(Nanowerk News) Researchers at Karolinska Institutet, Sweden, have developed a nanoparticle technology that can be used to stabilise membrane proteins so that their structure can be studied in a lipid environment. The method, described in Nature Methods ("A Saposin lipoprotein nanoparticle system for membrane proteins"), makes it possible to access drug targets that previously could not be investigated and therefore potentially allows for the development of novel drugs, therapeutic antibodies and vaccines.
|
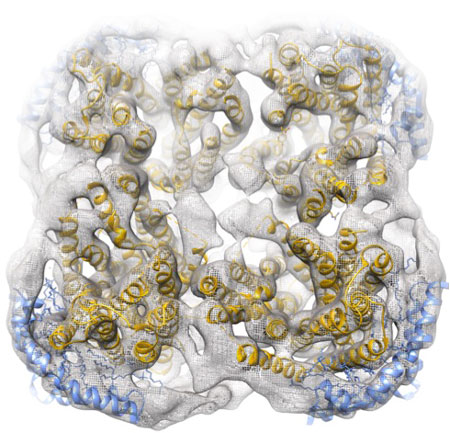 |
| An international research team have developed a nanoparticle technology that can be used to stabilise membrane proteins. (Image: Jens Frauenfeld et al.)
|
|
"Our technology, termed Salipro, may offer a wide range of potential applications, ranging from structural biology to the discovery of new pharmacological agents, as well as the therapeutic delivery of protein-based therapeutics and vaccines", says first author Jens Frauenfeld, who was working at the Department of Medical Biochemistry and Biophysics at Karolinska Institutet when the study was performed.
|
|
Membrane proteins are the targets of more than 60 per cent of drugs in clinical use. In addition, the membrane proteins of viruses are the key functional unit in commercial vaccines. Thus, membrane proteins are very important in biology, drug discovery and vaccination. The problem that researchers face is that these proteins are very unstable and therefore hard to investigate. They are embedded in membranes that are made up of different kinds of lipids. Most often, detergents are used to extract the membrane proteins. However, detergents are associated with protein instability and poor compatibility with structural and biophysical studies. Moreover, detergents do not provide a lipid environment, which is important for membrane proteins.
|
|
The investigators behind the new study worked around that problem by using the small cellular protein saposin. Usually, saposin shuttles lipids from one place to another within the cell. Since saposin is known to bind to lipids, the researchers evaluated whether it would be possible to develop a method to make stable saposin-based lipid nanoparticles. They then expanded the method so that it is possible to also embed fragile membrane proteins into those lipid nanoparticles and stabilise them.
|
|
The investigators demonstrate that the method facilitates high-resolution 3-dimensional studies of membrane proteins by single-particle electron cryo-microscopy, cryo-EM, an increasingly popular technique among scientists who want to study proteins at atomic resolution. They also present a method to extract and stabilise fragile membrane proteins from the HIV virus membrane.
|
|
"To our knowledge, the HIV spike protein preparation presented in the study using the Salipro system represents the first approach that allows the stabilisation of the HIV-1 spike, including the important membrane domains, in a soluble and functional state", says Professor Pär Nordlund at the Department of Oncology-Pathology.
|
|
The authors believe that the technology may also be applicable to other viral envelope proteins such as influenza virus haemagglutinin, ebola virus G-protein or hepatitis C virus E protein. Taken together, the researchers apply the method on three different membrane protein targets for structural and functional studies.
|

