| Posted: Oct 07, 2016 |
New, carbon-nanotube tool for ultra-sensitive virus detection and identification
(Nanowerk News) A new tool that uses a forest-like array of vertically-aligned carbon nanotubes that can be finely tuned to selectively trap viruses by their size can increase the detection threshold for viruses and speed the process of identifying newly-emerging viruses. The research, by an interdisciplinary team of scientists at Penn State, is published in the October 7, 2016 edition of the journal Science Advances ("Tunable and label-free virus enrichment for ultrasensitive virus detection using carbon nanotube arrays").
|
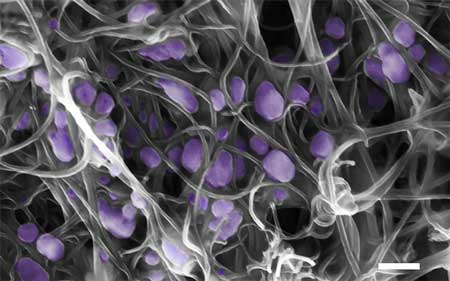 |
| Scanning electron microscope image (scale bar, 200 nm) of the H5N2 avian influenza virus (purple) trapped inside the aligned carbon nanotubes. (Image: Penn State University)
|
|
"Detecting viruses early in an infection before symptoms appear, or from field samples, is difficult because the concentration of the viruses could be very low -- often below the threshold of current detection methods," said Mauricio Terrones, professor of physics, chemistry, and materials science and engineering at Penn State, and one of the corresponding authors of the research.
|
|
"Early detection is important because a virus can begin to spread before we have the ability to detect it. The device we have developed allows us to selectively trap and concentrate viruses by their size -- smaller than human cells and bacteria, but larger than most proteins and other macromolecules -- in incredibly dilute samples. It further increases our ability to detect small amounts of a virus by more than a hundred times."
|
|
The research team developed and tested a small, portable device that increases the sensitivity of virus detection by trapping and concentrating viruses in an array of carbon nanotubes. Dilute samples collected from patients or the environment are passed through a filter to remove large particles such as bacteria and human cells, then through the array of carbon nanotubes in the device. Viruses get trapped and build up to usable concentrations within the forest of nanotubes, while other smaller particles pass through and are eliminated. The concentrated virus captured in the device can then be put through a panel of tests to identify it, including molecular diagnosis by polymerase chain reaction (PCR), immunological methods, virus isolation, and genome sequencing.
|
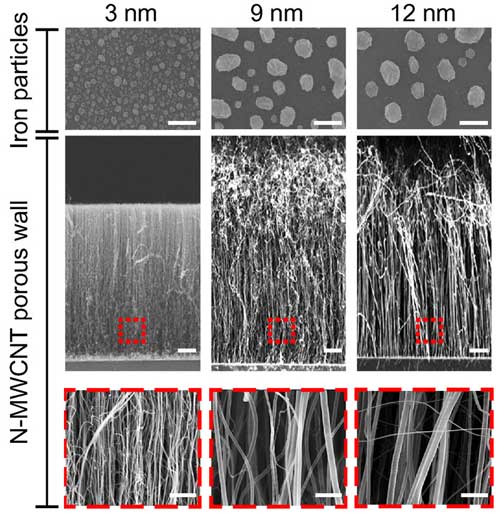 |
| Tunable inter-tubular distance of carbon nanotubes for matching virus dimensions. Scale bars, top: 100 nm, middle: 10 µm, bottom: 200 nm. (Image: Penn State University) (click on image to enlarge)
|
|
"Because our device isolates and concentrates viruses purely by size, we can capture viruses that we don't know anything about biologically -- we don't need any antibody or other molecular label," said Terrones. "Once we capture and concentrate the virus, we can then use other techniques such as whole-genome sequencing to characterize it."
|
|
"Most lethal viral outbreaks in the past two decades were caused by newly emerging viruses. This size-based virus-enrichment technology can be particularly powerful in identification of emerging viruses and discovery of new viruses that do not have antibodies and sequence information available," said Si-Yang Zheng, associate professor of biomedical engineering at Penn State, the other corresponding author on the paper. "Not only does our new technology enrich viruses by at least one hundred times, but it also removes host and environmental contaminants, and enables direct virus identification by next-generation sequencing from field-collected samples without virus culture."
|
|
Viruses -- such as influenza, HIV/AIDS, Ebola, and Zika -- can cause sudden, unpredictable outbreaks that lead to severe public-health crises. Currently available techniques for isolating and identifying the viruses that cause these outbreaks are slow, expensive, and use equipment and reagents that can be expensive, bulky, and require specialized storage. Additionally, many recent outbreaks have been caused by newly emerging viruses for which there are no established ways to selectively isolate them for identification and characterization.
|
 |
| Illustration of size-based virus enrichment by the aligned carbon nanotube array. (Image: Penn State University)
|
|
"We developed the technology to grow a forest of nanotubes and we can control the distance between the trunks," said Zheng. "The intertube distance can range from about 17 nanometers to over 300 nanometers to selectively capture viruses. The unique properties of the carbon-nanotube forest allow us to integrate it into a robust, scalable, and portable microdevice that can be adapted for use in the field without the need for bulky instruments and specialized storage of reagents."
|
|
The researchers validated the ability of their newly developed device to capture viruses from dilute samples using known concentrations of previously identified viruses as well as field samples of emerging and unknown viruses. "We developed a portable platform to enrich and isolate viruses based on their physical sizes," said Yin-Ting Yeh, a postdoctoral researcher at Penn State and first author of the paper. "This tunable size-based approach provides rapid virus enrichment directly from field samples without the use of antibodies. The device enables early detection of emerging diseases and potentially allows for vaccine development much sooner in the process of an outbreak."
|



