| Posted: Nov 16, 2016 |
The power of three
(Nanowerk News) Palladium-catalyzed organic reactions, such as Sonogashira cross-coupling, may be made more efficient and substrate-tolerant as a result of new findings at A*STAR.
|
|
Having previously developed a catalyst that outperforms existing state-of-the art analogs, the researchers now show how a catalyst structure sparks unprecedented activity (Organometallics, "Mechanistic insights and implications of dearomative rearrangement in copper-free Sonogashira cross-coupling catalyzed by Pd-Cy*Phine").
|
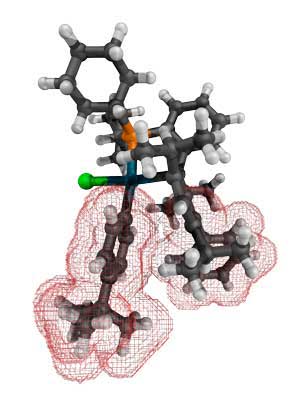 |
| Cy*Phine-based palladium catalyst for copper-free Sonogashira cross-coupling reactions. The benzene rings (highlighted in red) block potential side reactions involving the ligand. (Image: A*STAR Institute of Chemical and Engineering Sciences, A*STAR Institute of High Performance Computing, and A*STAR Singapore Bioimaging Consortium)
|
|
Sonogashira cross-coupling — which assembles molecules by joining terminal alkynes to chlorinated aromatic compounds, while keeping the triple bond intact — is used for synthesizing organic products destined to become pharmaceuticals and molecular electronics. Typically the assembling relies on a copper salt that increases the catalytic productivity by assisting the alkyne addition to the catalyst. However, this salt also promotes the formation of by-products which require complex and time-consuming purification.
|
|
Yee Hwee Lim and coworkers, from the A*STAR Institute of Chemical and Engineering Sciences and Singapore Bioimaging Consortium, have solved this issue by creating a palladium complex, without copper, that can still catalyze Sonogashira cross-coupling. Lim explains the less metal used, the better it is for downstream purification stages. “To use this catalyst in an industrial process, we want to decrease the metal content,” she adds.
|
|
As well as being more efficient, the copper-free system can be applied to a broader range of substrates than its commercially-available analogs. This performance enhancement hinges on the Cy*Phine ligand, which consists of a phosphine-type molecule bearing three interconnected benzene rings. However, its underlying mechanism remained unclear.
|
|
With Adrian Matthew Mak, from the A*STAR Institute of High Performance Computing, Lim’s team has computationally identified the main steps of the catalytic cycle. They discovered that the rate limiting step is where the alkyne binds to the catalyst–aromatic substrate complex.
|
|
When the catalytic activities achieved via Cy*Phine and a two-ring ligand were further compared, the team showed that during this alkyne addition, the third benzene ring blocked unwanted reactions (see image).
The A*STAR researchers who developed the palladium catalyst and investigated the role of its ligand in its performance.
|
|
“Without the third ring, a part of the catalyst could actually go through these unproductive pathways” says Lim “and this reduces the efficiency of the main catalytic cycle.” Explaining that side reactions proceed depending on their energy difference from the next catalytic step, she adds that, in contrast to the Cy*Phine-based catalyst, “these two energies are quite similar for the two-ring system.”
|
|
The Cy*Phine-based catalyst has been commercialized by Aspira Scientific for the past year. The team has applied the catalyst to different reactions and observed enhancement effects. Now, says Lim, “we are planning to look more closely at how it is affecting the catalytic cycle”.
|

