| Posted: Dec 23, 2016 |
Fluorescence microscopy: It cannot get any sharper
(Nanowerk News) It is the holy grail of light microscopy: improving the resolving power of this method such that one can individually discern molecules that are very close to each other.
|
|
Scientists around the Nobel laureate Stefan Hell at the Max Planck Institute for Biophysical Chemistry in Göttingen have now achieved what was for a long time considered impossible – they have developed a new fluorescence microscope, called MINFLUX, allowing, for the first time, to optically separate molecules, which are only nanometers (one millionth of a millimeter) apart from each other (Science, "Nanometer resolution imaging and tracking of fluorescent molecules with minimal photon fluxes").
|
|
This microscope is more than 100 times sharper than conventional light microscopy and surpasses even the best super-resolution light microscopy methods to date, namely STED developed by Hell and PALM/STORM described by Nobel laureate Eric Betzig, by up to 20 times. For MINFLUX, Hell used the advantages of STED and PALM/STORM in a completely new concept. This breakthrough opens up new opportunities for researchers to investigate how life functions at the molecular level.
|
 |
| With MINFLUX microscopy one can, for the first time, separate molecules optically which are only a few nanometers apart from each other. On the left, a schematic of the fluorescing molecules is presented. Whereas the ultra-high resolution PALM/STORM microscopy at the same molecular brightness (right) delivers a diffuse image of the molecules (here in a simulation under ideal technical conditions), the position of the individual molecules can be easily discerned with the practically realized MINFLUX (middle). (Image: MPI f. Biophysical Chemistry/ K. Gwosch )
|
|
“We have routinely achieved resolutions of a nanometer with MINFLUX, which is the diameter of individual molecules – the ultimate limit of what is possible in fluorescence microscopy,” explains Hell, Director at the Max Planck Institute for Biophysical Chemistry. “I am convinced that MINFLUX microscopes have the potential to become one of the most fundamental tools of cell biology. With this concept it will be possible to map cells in molecular detail and to observe the rapid processes in their interior in real time. This could revolutionize our knowledge of the molecular processes occurring in living cells.”
|
|
The Göttingen physicist, who also works at the Max Planck Institute for Medical Research and the German Cancer Research Center in Heidelberg, has long been convinced that fluorescence microscopy resolution can be increased down to the dimension of individual molecules – with classical use of focused light and conventional lenses.
|
|
In fact, the physicist Ernst Abbe had formulated in 1873 that the resolution of light microscopes is limited to half the wavelength of light, which is about 200 nanometers. More than 100 years later, this Abbe limit is still valid. However, Hell was the first to show that this limit can be overcome with STED microscopy, which he conceived in 1994 and established experimentally five years later.
|
|
STED as well as PALM/STORM, developed a few years later, in practice achieve a separation sharpness of about 20 to 30 nanometers – about ten times better than the Abbe limit. For the development of these ultra-high resolution light microscopy techniques, Hell and Betzig together with William E. Moerner were awarded the 2014 Nobel Prize in Chemistry.
|
Advantages of STED and PALM/STORM combined
|
|
Both STED and PALM/STORM separate neighboring fluorescing molecules by switching them on and off one after the other so that they emit fluorescence sequentially.
|
|
However, the methods differ in one essential point: STED microscopy uses a doughnut-shaped laser beam to turn off molecular fluorescence at a fixed location in the sample, i.e. everywhere in the focal region except at the doughnut center.
|
|
The advantage is that the doughnut beam defines exactly at which point in space the corresponding glowing molecule is located. The disadvantage is that in practice the laser beam is not strong enough to confine the emission to a single molecule at the doughnut center.
|
|
In the case of PALM/STORM, on the other hand, the switching on and off is at random locations and at the single-molecule level. The advantage here is that one is already working at the single-molecule level, but a downside is that one does not know the exact molecule positions in space. The positions have to be found out by collecting as many fluorescence photons as possible on a camera; more than 50,000 detected photons are needed to attain a resolution of less than 10 nanometers.
|
|
In practice, one therefore cannot routinely achieve molecular (one nanometer) resolution.
|
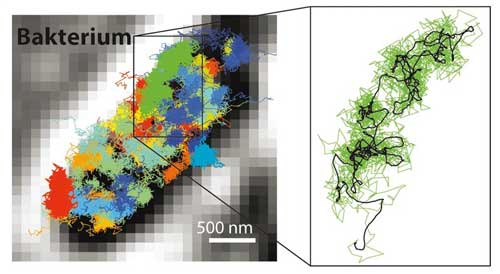 |
| With MINFLUX it is possible to follow many much faster movements than possible with STED or PALM/STORM microscopy. It is therefore possible to make the movements of fluorescence labeled molecules visible in a living cell. Left: Movement pattern of 30S ribosomes (parts of protein factories, colored) in an E. coli bacterium (black-white). Right: Movement pattern of a single 30S ribosome (green) shown enlarged. (Image: MPI f. Biophysical Chemistry/ Y. Eilers ) (click on image to enlarge)
|
|
Hell had the idea to uniquely combine the strengths of both methods in a new concept. “This task was anything but trivial. But my co-workers Francisco Balzarotti, Yvan Eilers, and Klaus Gwosch have done a wonderful job in implementing this idea experimentally with me.” Their new technique, called MINFLUX (MINimal emission FLUXes), is now introduced by Hell together with the three junior scientists as first authors in Science.
|
|
MINFLUX, like PALM/STORM, switches individual molecules randomly on and off. However, at the same time, their exact positions are determined with a doughnut-shaped laser beam as in STED. In contrast to STED, the doughnut beam here excites the fluorescence.
|
|
If the molecule is on the ring, it will glow; if it is exactly at the dark center, it will not glow but one has found its exact position. Balzarotti developed a clever algorithm so that this position could be located very fast and with high precision.
|
|
“With this algorithm it was possible to exploit the potential of the doughnut excitation beam,” the young scientist explains. Gwosch, who obtained the molecular resolved images, adds “It was an incredible feeling as we, for the first time, were able to distinguish details with MINFLUX on the scale of a few nanometers.”
|
100 times better resolution
|
|
In addition to the molecular resolution, the combination of STED and PALM/STORM offers an additional major advantage: “MINFLUX is much faster in comparison. Since it works with a doughnut laser beam, it requires much lower light signal, i.e. fewer fluorescence photons, per molecule as compared to PALM/STORM for attaining the ultimate resolution,” Hell states.
|
|
Already with STED one could record real-time videos from the inside of living cells. But now it was possible to trace the movement of molecules in a cell with a 100 times better temporal resolution, as Eilers emphasizes. He managed to film the movement of molecules in a living E. coli bacterium with MINFLUX for the first time, with an unprecedented spatio-temporal resolution.
|
|
“As far as speed is concerned, we have not made the most of the possibilities with MINFLUX,” Eilers says.
|
|
The researchers are convinced that even extremely fast-occurring changes in living cells can be investigated in the future, like for example the movement of cellular nanomachines or the folding of proteins.
|


