| Posted: Feb 28, 2018 |
How exact is my nanoruler?
(Nanowerk News) The development and evaluation of DNA origami-based nanorulers enables measurements in the nano cosmos with increasing precision. Nanosystems Initiative Munich (NIM) scientist Prof Dr Philip Tinnefeld and his team explore such self-assembled nano structures.
|
|
To measure distances in our daily life, normally we utilze rulers or a folding ruler. Quantification takes place by comparing the object or distance and the ruler. But how can we measure distances on the molecular scale and standardize our measurements?
|
|
Under the guidance of Professor Philip Tinnefeld, researchers at the LMU Munich and the TU Braunschweig developed nanorulers with fluorescent markers in precisely determined distances along a DNA nanostructure. Special feature is the highly parallelized self-assembly of the DNA origami-based nanorulers.
|
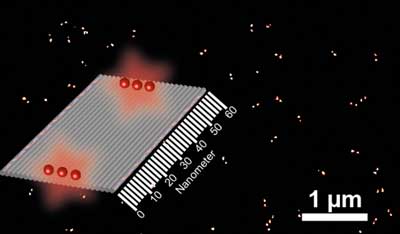 |
| Schematic of a nano ruler with DNA-coupled fluorescent dyes (red) on a DNA origami square (grey). The bright double signal on the black background presents highly resolved images of nanostructures acquired by super-resolution fluorescence microscopy. (Image: M Raab)
|
|
Potential applications are various, the worldwide first commercial utilization of DNA origami are such nanorulers usable as reference structures for the calibration of microscopes and as positive controls in super-resolution fluorescence microscopy.
|
|
From the metrological perspective, there is the question about accuracy and precision of DNA origami-based nanorulers. Are such self-assembled nanorulers capable for the quantitative characterization of microscale properties, and how precise is the distance determination?
|
|
Those and other metrologic questions, for example the traceability of measured data to the SI units are the main focus of the National Metrology Institute of Germany (PTB) in Braunschweig. This common research interest led to a collaboration of professor Tinnefeld (LMU) and scientists of the TU Braunschweig and the PTB (Departments of Imaging and Wave Optics, and Precision Engineering). Their results are published in Scientific Reports ("Using DNA origami nanorulers as traceable distance measurement standards and nanoscopic benchmark structures").
|
DNA origami-based nanorulers
|
|
The distance between two fluorescent marks on DNA origami-based nanorulers can be determined with an uncertainty of only 1-2 nm. It has to be highlighted that the reproducibility of the measurements over several days and on different microscopes was excellent. “The deviation of determined distances was mostly below one nanometer!” stresses Mario Raab, first author of the study. “This calibration technique allows to use our nanorulers not only as samples but also as standards.”
|
|
“The accuracy and reproducibility we reach with these self-assembled structures are amazing. Crystalline structures with countable atom layers often serve as standards in nanosciences. These DNA origami-based nanorulers open a new path to broadly available reference structures in a simple and elegant way.”, explain the partners at PTB.
|
|
Biophysical measurement techniques like the super-resolution microscopy were developed in recent years in a broad variety but with limited standardization. Numerous potential applications lay in the medical field. If such devices some day will decide on healthy or not, their reliability has to be ensured quantitatively. This work provides an approach to determine uncertainties and minimize them.
|
Nanorulers in expasion microscopy
|
|
In their latest work ("Quantifying Expansion Microscopy with DNA Origami Expansion Nanorulers"), Dr Max Scheible and professor Philip Tinnefeld describe the application of DNA origami-based nanorulers in a new microscopy approach, the expansion microscopy (ExM).
|
|
There, the sample is physically expanded to increase its size by a multiple and therefore allow imaging by conventional fluorescence microscopy as an alternative to super-resolution microscopy. The sample is embedded in an electrolytic polymer and labeled with an antibody-fluorophore complex before the gel gets expanded. Then the original sample gets digested, but the expanded gel mimicks its shape, structure and dimension in 3-5-fold magnification. The DNA origami-based nanorulers labeled with fluorophores enable calibration and quantification of the ExM.
|
Optical voltage sensing nano-devices
|
|
Philip Tinnefeld and colleagues present the proof-of-principle for new optical voltage sensing nano-devices based on voltage-responsive DNA origami labeled with FRET dyes (Nano Letters, "Optical Voltage Sensing Using DNA Origami"). DNA origami serves basis for the positioning of such fluorescent dyes with nanometer precision.
|
|
They could show that an increase in voltage induces gradual distortions as well as distinct changes in DNA conformation. These unique features enables the design of dynamic voltage-responsive molecular rulers with optical read out. In simulations and experiments they demonstrate the successful use of a DNA origami structure labeled with a FRET-dye pair, whose FRET signal is calibrated in response to the applied potential.
|
Virus detection using optical nanoantennas
|
|
In another approach, the group of Professor Tinnefeld developed optical nanoantennas for the detection of Zika virus DNA or RNA in both, buffer and heat-inactivated human blood serum (Analytical Chemistry, "Optical Nanoantenna for Single Molecule-Based Detection of Zika Virus Nucleic Acids without Molecular Multiplication"). They possess high sensitivity against small nucleotide variations, allowing discrimination of closely related pathogens.
|
|
The 125 nm DNA origami-based optical antennas carry complementary nucleic acid strands and upon binding of the target sequence from the sample, there is a fluorescence signal. That fluorescence signal is enhanced as the antennas utilize metallic nanoparticles to create a hotspot. Aim is the application in single-molecule-based point of care diagnosis. The incorporation of orthogonal fluorescent labels allowed for multiplexing, the simultaneous detection of different sequences.
|
Hybrid structures for fluorescence enhancement
|
|
Coupling of a natural light-harvesting complex, peridinin-chlorophyll a-protein, to dimer DNA origami-based optical antennas results in Opens external link in new windowhybrid constructs. These systems also utilize colloidal gold and silver nanoparticles at the hotspots of the optical nanoantennas. In selective excitation of the different pigments in the harvesting complex, 500-fold fluorescence enhancement was reached.
|

