| Posted: Oct 18, 2018 | |
Nanocages in the lab and in the computer: How DNA-based dendrimers transport nanoparticles(Nanowerk News) How to create nanocages, i.e., robust and stable objects with regular voids and tunable properties? Short segments of DNA molecules are perfect candidates for the controllable design of novel complex structures. |
|
| Physicists from the University of Vienna, the Technical University of Vienna, the Foschungszentrum Jülich in Germany and Cornell University in the U.S.A., investigated methodologies to synthesize DNA-based dendrimers in the lab and to predict their behavior using detailed computer simulations. Their results are published in Nanoscale ("Structure and stimuli-responsiveness of all-DNA dendrimers: theory and experiment"). | |
 |
|
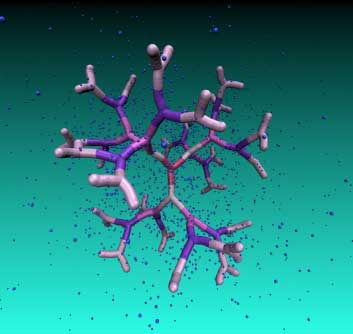
| DNA-based dendrimer of the fifth generation immersed in a counterion solution. Regular voids in the interior of this novel macromolecule can be employed as nano-carriers. (Image: Natasa Adzic) | |
| Nanocages are highly interesting molecular constructs, from the point of view of both fundamental science and possible applications. The cavities of these nanometer-sized objects can be employed as carriers of smaller molecules, which is of critical importance in medicine for drug or gene delivery in living organisms. | |
| This idea brought together researchers from various interdisciplinary fields who have been investigating dendrimers as promising candidates for creating such nano-carriers. Their tree-like architecture and step-wise growth with repeating self-similar units results in dendrimers containing cavities, hollow objects with controllable design. | |
| Nevertheless, decades of research have showed that vast number of different dendrimer types experience back-folding of outer branches with growing dendrimer generations, giving rise to a higher density of constituents in the molecule's interior. The effect of back-folding is enhanced upon addition of salt in the solution, whereby flexible dendrimers undergo significant shrinking, becoming compact objects with no hollow spaces in their interior. | |
| The team of collaborators consisted of Natasa Adzic and Christos Likos (University of Vienna), Clemens Jochum and Gerhard Kahl (TU Vienna), Emmanuel Stiakakis (Jülich) as well as Thomas Derrien and Dan Luo (Cornell). The researchers found a way to create dendrimers rigid enough to prevent back-folding of outer arms even in the case of high branching generations, preserving regular voids in their interior. | |
| Moreover, their novel macromolecules are characterized by remarkable resistance to added salt: they showed that the morphology and conformational characteristics of these systems stay unaffected even upon of addition of salt even at high concentration. The nanocages they created, in the lab and studied computationally are DNA-based dendrimers, or so-called, dendrimer-like DNAs (DL-DNA). | |
| The building block they are composed of is a Y-shaped double-stranded DNA unit, a three-armed structure consisting of double-stranded DNA (ds-DNA), formed via hybridization of three single-stranded DNA chains (ss-DNA), each of which has partially complementary sequences to the other two. Each arm is made up of 13 base pairs and a single-stranded sticky end with four nucleobases which acts as a glue. | |
| While a single Y-DNA corresponds to the first dendrimer generation, the attachment of further Y-DNA elements yields DL-DNA of higher generations. The resulting dendrimer is a charged and hollow-containing macromolecular assembly with tree-like architecture. Due to the rigidity of dsDNA, the branches of DL-DNA are stiff so that the whole molecule is rigid. Since DNA is charged, the electrostatic repulsion enhances the rigidity of the molecule. | |
| DL-DNA molecules have been assembled in the laboratory by the Jülich and Cornell partners with remarkable control and sub-nanometer precision through programmable sticky-end cohesions. Their step-wise growth is highly controllable, unidirectional and non-reversible. | |
| This property is of high importance, as it has been shown that DNA-based dendrimers have been envisioned to play a promising role in developing nanoscale-barcodes, DNA-based vaccine technologies, as well as a structural probes involving multiplexed molecular sensing processes. Sizes, shapes as well as additional conformational details invisible to the experimentalists, such as the size of voids and the degree of branches back-folding, have been analyzed by computer simulations in Vienna. | |
| To describe the complex structure of DNA units, the group used a simple monomer-resolved model with interactions carefully chosen to mimic the equilibrium properties of DNA in physiological solution. The excellent agreement obtained between experiments and simulations for the dendrimer characteristics validates the theoretical models employed and paves the way for further investigation of the nanocages’ properties and their applications as functional and smart nanocarriers and as building blocks for engineering biocompatible artificial materials. |
| Source: University of Vienna | |
|
Subscribe to a free copy of one of our daily Nanowerk Newsletter Email Digests with a compilation of all of the day's news. |
