| Oct 19, 2023 |
Researchers design a pulsing nanomotor
(Nanowerk News) An international team of scientists headed by the University of Bonn has developed a novel type of nanomotor. It is driven by a clever mechanism and can perform pulsing movements. The researchers are now planning to fit it with a coupling and install it as a drive in complex machines.
|
Key Takeaways
|
|
Researchers have developed a new type of nanomotor that performs pulsing movements similar to a hand grip trainer, but is a million times smaller.
The nanomotor uses RNA polymerases to move along a DNA strand, pulling its handles closer together in a cycle, mimicking the function of proteins in cells.
This unique motor is powered by nucleotide triphosphates, the same energy source used by cells to create proteins.
The motor has been demonstrated to be easily combinable with other structures, suggesting its potential use in complex nanomachines.
Further work is being done to optimize the nanomotor's performance, including the development of a clutch system to control its activity.
|
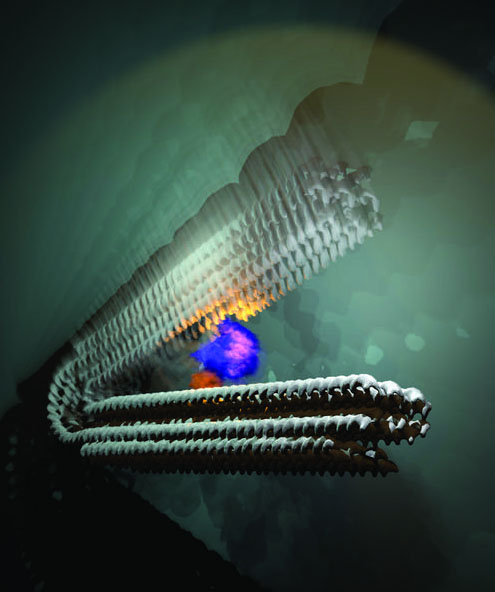 |
| The novel type of nanomotor with an RNA polymerase, which pulls the two “handles” together and then releases them again. This generates a pulsing movement. (Image: Mathias Centola, University of Bonn)
|
The Research
|
|
The team's findings have now appeared in the journal Nature Nanotechnology ("A rhythmically pulsing leaf-spring DNA-origami nanoengine that drives a passive follower").
|
|
This novel type of motor is similar to a hand grip trainer that strengthens your grip when used regularly. However, the motor is around one million times smaller. Two handles are connected by a spring in a V-shaped structure.
|
|
In a hand grip trainer, you squeeze the handles together against the resistance of the spring. Once you release your grip, the spring pushes the handles back to their original position. “Our motor uses a very similar principle,” explains Prof. Dr. Michael Famulok from the Life and Medical Sciences (LIMES) Institute at the University of Bonn. “But the handles are not pressed together but rather pulled together.”
|
|
For this purpose, the researchers have repurposed a mechanism without which there would be no plants or animals. Every cell is equipped with a sort of library. It contains the blueprints for all types of proteins that the cell needs to perform its function. If the cell wants to produce a certain type of protein, it orders a copy of the respective blueprint. This transcript is produced by RNA polymerases.
|
RNA polymerases drive the pulsing movements
|
|
The original blueprint consists of long strands of DNA. The RNA polymerases move along these strands and copy the stored information letter by letter. “We took an RNA polymerase and attached it to one of the handles in our nanomachine,” explains Famulok, who is also a member of the transdisciplinary research areas “Life & Health” and “Matter” at the University of Bonn. “In close proximity, we also strained a DNA strand between the two handles. The polymerase grabs on to this strand to copy it. It pulls itself along the stand and the non-transcribed section becomes increasingly smaller. This pulls the second handle bit by bit towards the first one, compressing the spring at the same time.”
|
|
The DNA strand between the handles contains a particular sequence of letters shortly before its end. This so-called termination sequence signals to the polymerase that it should let go of the DNA. The spring can now relax again and moves the handles apart. This brings the start sequence of the strand near to the polymerase and the molecular copier can start a new transcription process: The cycle thus repeats.
|
|
“In this way, our nanomotor performs a pulsing action,” explains Mathias Centola from the research group headed by Prof. Famulok, who carried out a large proportion of the experiments.
|
An alphabet soup serves as fuel
|
|
This motor also needs energy just like any other type of motor. It is provided by the “alphabet soup” from which the polymerase produces the transcripts. Every one of these letters (in technical terminology: nucleotides) has a small tail consisting of three phosphate groups – a triphosphate. In order to attach a new letter to an existing sentence, the polymerase has to remove two of these phosphate groups. This releases energy which it can use for linking the letters together.
|
|
“Our motor thus uses nucleotide triphosphates as fuel,” says Famulok. “It can only continue to run when a sufficient number of them are available.”
|
|
By monitoring individual nanomotors, one of the cooperation partners based in the US state of Michigan was able to demonstrate that they actually carry out the expected movement. A research group in Arizona also simulated the process on high speed computers. The results could be used, for example, to optimize the motor to work at a particular pulsation rate.
|
|
Furthermore, the researchers were able to demonstrate that the motor can be easily combined with other structures. This should make it possible for it to, for example, wander across a surface – similar to an inchworm that pulls itself along a branch in its own characteristic style.
|
|
“We are also planning to produce a type of clutch that will allow us to only utilize the power of the motor at certain times and otherwise leave it to idle,” explains Famulok. In the long term, the motor could become the heart of a complex nanomachine. “However, there is still a lot of work to be done before we reach this stage.”
|

