| Posted: November 16, 2006 |
Nanotechnology tools yield DNA transcription breakthrough |
|
(Nanowerk News) Rutgers researcher Richard H. Ebright and his collaborators have resolved key questions regarding transcription, the fundamental life process that was the subject of the 2006 Nobel Prize in Chemistry.
|
|
Transcription is the first step in the process cells employ to read and carry out the out instructions contained in genes. Transcription is carried out by a molecular machine known as RNA polymerase, which synthesizes an RNA copy of the information in DNA.
|
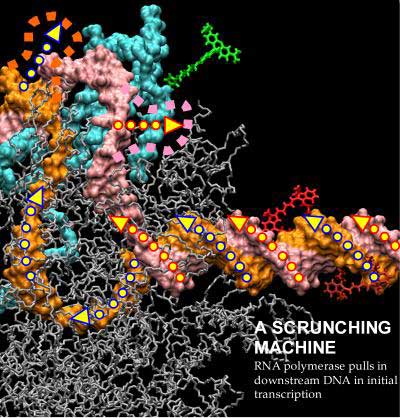 |
| Initial transcription by RNA polymerase proceeds through a "DNA scrunching" mechanism, in which the enzyme remains stationary on promoter DNA and pulls into itself downstream DNA. [Proposed movements of the template and nontemplate DNA strands are indicated by blue outlined and red-outlined arrows. Proposed positions at which the scrunched template and nontemplate DNA strands emerge from the enzyme are indicated by orange and pink dashed lines. Positions of fluorescent probes used to analyze scrunching are indicated in green (donor probe on polymerase), brick red (acceptor probe at promoter position +20 in absence of scrunching), and bright red (acceptor probe at promoter position +20 in presence of scrunching).] (Image provided by Achillefs Kapanidis, Shimon Weiss, and Richard H. Ebright.)
|
|
Two papers by Ebright and collaborators in the Nov. 17 issue of the journal Science define for the first time the mechanisms by which the machine begins synthesis of RNA and then breaks free from its initial binding site to move along DNA to continue synthesizing RNA.
|
|
The results establish that during transcription initiation the machine remains stationary at its initial binding site and "reels in" adjacent DNA segments, unwinding these segments and pulling the unwound DNA strands into itself. This remarkable mechanism, termed "DNA scrunching," enables the machine to acquire and accumulate the energy it needs to break its binding interactions with the initial binding site, and to begin to move down the gene.
|
|
"Our findings were made possible by newly developed, single-molecule methods," said Ebright, a Howard Hughes Medical Institute investigator and professor of chemistry and chemical biology at Rutgers, The State University of New Jersey. "These methods enabled us to analyze and manipulate individual molecules of the machine, one-by-one, as they carried out reactions."
|
|
The discoveries significantly advance our understanding of the structure and function of the molecular machine that carries out transcription, setting the stage for new opportunities in combating the bacterial diseases that kill 13 million persons each year worldwide.
|
|
"For six decades, antibiotics have been our bulwark against bacterial infectious diseases, but this bulwark now is collapsing," said Ebright. "For all major bacterial pathogens, including tuberculosis, strains such as XDR-TB that are resistant to current antibiotics have emerged."
|
|
In addition, bacterial pathogens that may be used in bioterrorism can be engineered, and, in the former Soviet Union, were intentionally engineered to be resistant to current antibiotics.
|
|
Ebright explained that his laboratory at Rutgers' Waksman Institute of Microbiology has two parts: One part seeks a fundamental understanding of the machine; the other uses that understanding to develop new classes of antibacterial agents that function by inhibiting the bacterial version of the machine. "There is a direct information flow from our basic research to our applied research," he said. "Our basic research identifies new vulnerabilities within the bacterial version of the machine; our applied research exploits those vulnerabilities."
|
|
One of the studies reported in Science was conducted by Ebright's laboratory in conjunction with Shimon Weiss' laboratory at the California NanoSystems Institute of the University of California-Los Angeles (UCLA). The study by the Rutgers/UCLA team used single-molecule fluorescence spectroscopy. The researchers attached pairs of fluorescent "tags" to key structural elements of the machine and then monitored changes in distance between tags in single molecules as transcription occurred. The researchers showed that, during initial transcription, the machine does not move to reach adjacent DNA segments; nor does it stretch to reach adjacent DNA segments (as had been proposed two decades ago in models termed "transient excursions" and "inchworming"). Instead, the researchers showed that the machine remains stationary and pulls adjacent DNA segments into itself.
|
|
"The study of molecular machines and the dynamics of their moving parts hold great interest for nanotechnologists," said Weiss, leader of the UCLA group. "Beyond furthering the understanding of transcription, the novel methods and findings of this work will aid future studies of other molecular machines involved in cell replication, transcription and protein synthesis."
|
|
The other study reported in Science was conducted by Ebright's laboratory in collaboration with Terence Strick's laboratory at the Institut Jacques Monod in Paris. The study by the Rutgers/Paris team used single-molecule nanomanipulation. The researchers used an instrument referred to as "magnetic tweezers" to hold, stretch and twist a single molecule of DNA having a single start site for transcription. They then read out changes in the conformation of the DNA molecule in real time as transcription occurred. The researchers showed that the molecular machine responsible for transcription unwinds adjacent DNA segments and pulls unwound DNA into itself during initial transcription ("scrunching").
|
|
In addition, the researchers showed that machine rewinds this unwound DNA when the machine leaves the start site and begins to move down the gene ("unscrunching"). Finally, the researchers showed that this process of scrunching and unscrunching occurs every time that transcription initiation occurs, indicating that the process is an obligatory part of transcription initiation.
|
|
Taken together, the two studies answer the longstanding question of how the machine acquires the energy required to break its interactions with, and leave, the start site. The machine acquires this energy by unwinding DNA and pulling unwound DNA during initial transcription. As DNA is unwound, energy is stored in the system, in the same manner, Ebright notes, as winding the rubber band of a rubber-band-powered airplane stores energy. Eventually, there is sufficient energy stored in the system that the machine is able to break its interactions with the start site, to shoot forward and, at the same instant, to rewind the unwound DNA.
|
|
Ebright stated that the publication of these findings is part of a long-term collaboration with the UCLA and the Institut Jacques Monod groups. "Our ties are close," he said. "Three former Rutgers graduate students have gone on from my lab to the Weiss lab, and two other Rutgers graduate students have worked in the Strick lab during their thesis studies."
|

