| Posted: June 25, 2010 |
Illuminating the genetic alphabet |
|
(Nanowerk News) The development of fluorescent tags—small, light-emitting molecules attached to DNA strands—has revolutionized cell biology over the past two decades, allowing precise tracking of labeled segments in living systems. Because fluorescent tags project outwards from the nucleic acid chains, they can interfere with the mobility and geometry of their targets, skewing the view of critical biological actions.
|
|
Now, a research team led by Ichiro Hirao at the RIKEN Systems and Structural Biology Center in Yokohama has synthesized an unnatural fluorescent base pair system that can be incorporated directly into DNA and RNA molecules ("A Unique Fluorescent Base Analogue for the Expansion of the Genetic Alphabet"). Because this new addition to the genetic alphabet only minimally disturbs the delicate biochemical functions of cells, it holds great potential for advanced medical techniques such as DNA-based diagnostic testing.
|
|
Hirao and colleagues had previously discovered that a fluorescent 2-amino-6-thienylpurine molecule containing a thiophene ring, termed 's', formed a base pair with a pyrrole–aldehyde compound known as 'Pa' ("Fluorescent probing for RNA molecules by an unnatural base-pair system"). The new s–Pa pair could be site-specifically incorporated into RNA chains alongside the natural nucleotides; unfortunately, DNA replication processes with the s–Pa system were not as successful.
|
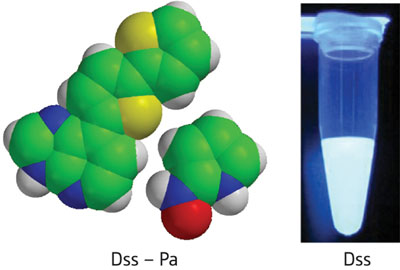 |
| Figure 1: New, unnatural base pairs called Dss–Pa (left) can be specifically incorporated into DNA and RNA molecules while retaining bright fluorescent emissions (right).
|
|
To solve this problem, the research team altered the structure of 's', removing the amino group and a nitrogen in the purine ring, then adding an extra thiophene ring to give a new nucleotide, called 'Dss.' This highly fluorescent molecule retained the same complimentary base pairing with Pa (Fig. 1), and could be easily transcribed into specific RNA positions by cell enzymes. Furthermore, single-nucleotide insertion experiments with Escherichia coli DNA polymerase I revealed that the Dss–Pa pairs could be replicated with nearly the same efficiency as natural bases.
|
|
Intriguingly, Dss also acted as a universal base—forming thermodynamically stable pairs with the four natural bases of DNA. Hirao says that the mechanisms of duplex DNA formation and polymerase reactions in replication and transcription with Dss are completely different than for natural systems.
|
|
Fluorescent imaging of functional DNA and RNA is becoming increasingly important as researchers seek to understand the conformations of these biopolymers in liquid environments. "The fluorescent intensity of the unnatural bases change greatly depending on the three-dimensional structure—providing a powerful tool to analyze the local structure of DNA and RNA molecules in solution," says Hirao.
|
|
Hirao envisages a wide range of basic and applied technologies arising from these bright new pieces of genetic code. "We are now applying Dss to molecular beacons, realtime PCR, and structural analysis," he says.
|

