| May 09, 2011 |
Graphene nanoribbons make the difference for fast nanochannel-based DNA sequencing
|
|
(Nanowerk News) DNA sequencing using current techniques involves chopping up DNA strands into small pieces, followed by amplification, transcription and finally optical identification of the constituent nucleotides. Before personalized medicine based on genetic screening can become a reality, however, this laborious and expensive process must be made simpler and faster.
|
|
Kwang Kim and co-workers at the Pohang University of Science and Technology in Korea have now proposed a model for a high-speed device that can sequence entire DNA strands using a nanochannel structure in combination with graphane nanoribbons ("Fast DNA sequencing with a graphene-based nanochannel device").
|
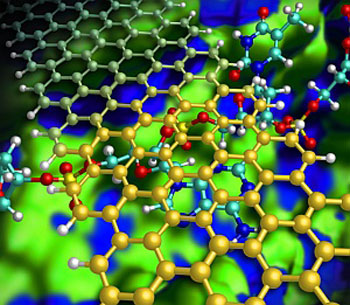 |
| Illustration showing how a graphene nanoribbon (gold) helps to keep a DNA strand (red/blue) flat as it passes through a nanochannel (green/blue).
|
|
The use of conductive nanoscale pores, just wide enough for single DNA strands, for sequencing has been studied for over 15 years. The concept is relatively straightforward: DNA passing through the pores produces a distinctive electrical signature corresponding to its nucleotide composition. Researchers hope to one day develop inexpensive sequencing microchips based on this mechanism, but so far the nanopore scheme has been unable to achieve accurate single nucleotide resolution due to inadequate control of DNA movement.
|
|
Kim and his team designed a hypothetical nanochannel device that radically improves sequencing accuracy by introducing graphene nanoribbons — narrow strips of carbon just one atom thick. In this model (see image), a DNA strand flows down a silicon nitride nanochannel and passes beneath a narrow graphene nanoribbon bridge. Because graphene and nucleotides share similar benzene-like rings, attractive aromatic stacking forces can force the DNA to lie flat in the channel, preventing the random movements of the DNA strand that have previously impeded performance.
|
|
The simulations also showed that graphene's unique electronic properties allow for highly sensitive nucleotide detection. Electrons usually move with zero resistance through the graphene framework, but in the presence of DNA strands, the conductance of the graphene nanoribbon dips sharply following a pattern specific to each nucleotide. The researchers developed a sophisticated algorithm that turned the time-dependent conductance of the device into a precise, automatic read-out of the DNA sequence.
|
|
Importantly, the proposed device can be built using existing clean-room protocols, meaning that it may soon be in the hands of researchers. "Identifying a single nucleotide on a graphene nanoribbon would be a significant first step in this direction," says Kim.
|

