| Aug 07, 2012 |
First DNA design guides available for GenoCon2 contest
|
|
(Nanowerk News) Manuals for designing a synthetic promoter using the DNA CAD environment for Challenge A, as well as pointers for designing DNA sequences to introduce plant functions for Challenge B are as follows:
|
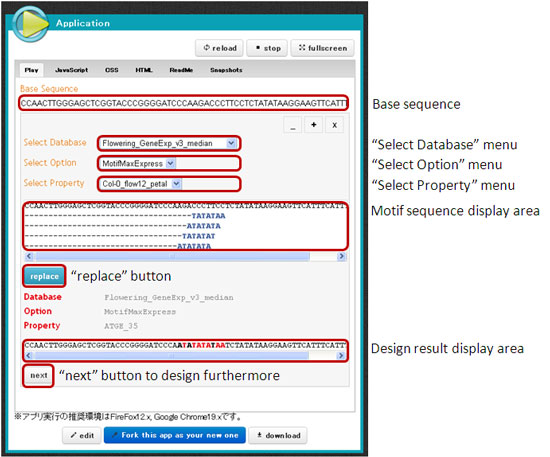 |
| Sample DNA design application.
|
|
Manuals for Challenge A
|
|
Challenge A – DNA design manual
|
|
Overview of the process to design a synthetic promoter for tissue-specific expression of firefly Luciferase in Arabidopsis plant body for Challenge A. This is a tutorial for creating an account on LinkData.org, loading the DNA CAD environment, mining functional sequences from the regulatory databases, and using them to design a synthetic promoter function. Includes an overview of the data structures for programmers.
|
|
Promoter Annotation Workflow
|
|
This is a tutorial for the process to find a promoter’s DNA sequence, annotate it with functional sequence elements, and upload the annotated sequence to LinkData.org so it can be accessed by the LinkDataApp.
|
|
Tips for Challenge B
|
|
Challenge B – Tips for Challenge B
|
|
Some pointers on the process to design DNA sequences by using those preceding research data showing that a plant gets a function to eliminate formaldehyde by inserting two enzymes, HPS (3-hexulose-6-phosphate synthase) and PHI (6-phospho-3-hexuloisomerase) into the Calvin Cycle of the plant.
|
|
About GenoCon2
|
|
The second international contest for rational genomic design, GenoCon2, will run this summer and fall. Hosted by the BASE (Bioinformatics And Systems Engineering) division at RIKEN, the contest will use a novel semantic software platform to modify the genome of the model flowering plant Arabidopsis thaliana.
|
|
A completely new Genocon2 challenge will ask participants to design plant promoter sequences with our DNA CAD (Computer Aided Design) environment, that uses cutting-edge gene regulatory databases to inform sequence design operations. These data have been semantically linked together and hosted on LinkData.org. GenoCon2 will also build upon the challenge from the previous iteration, to improve the design of synthetic plants for formaldehyde removal.
|
|
All Genocon2 contest activities are bioinformatic only: DNA design, sequence function programming with the App.LinkData.org system, and natural plant promoter annotation. Qualifying designs will be constructed into genetically modified plants, grown, and tested in a GenoCon2 laboratory, by expert researchers in a closed and safe environment.
|
|
For details of the challenges see the official website: genocon.org/
|

