| Oct 01, 2012 |
Researchers develop new method for detection of specific DNA sequences
|
|
(Nanowerk News) The detection of specific DNA sequences is central to the identification of disease-causing pathogens and genetic diseases, as well as other activities. But current detection technologies require amplification by polymerase chain reaction (PCR), fluorescent or enzymatic labels, and expensive instrumentation.
|
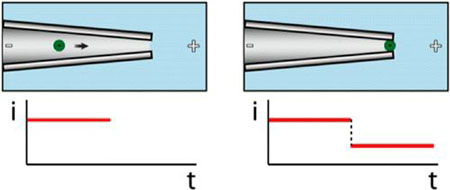 |
| In the presence of the DNA or RNA sequence of interest, beads are driven toward a sensing pore (left), which they block (right) producing a large signal easily readout with a handheld device. (Image: Jacob Schmidt)
|
|
Now, researchers from the UCLA Henry Samueli School of Engineering and Applied Science have developed a device that amplifies the size of a DNA molecule by coupling it to microscale beads that are one-hundredth the size of a human hair ("Sequence-specific Nucleic Acid Detection from Binary Pore Conductance Measurement").
|
|
In the presence of the specific DNA sequence to be detected, the beads will produce a large electronic signal that is easily read out by a simple device. This signal is generated because the binding of the specific DNA sequence to the beads gives them a negative charge, attracting them to a small pore, which they block. The electrical measurement of this blockage can be indicated by a simple light-emitting diode in a compact handheld device.
|
|
Impact
|
|
The simplicity and compactness of this new technology may enable low-cost handheld devices for DNA detection without the need for molecular amplification or any additional instrumentation. The method could be used in screening for genetic diseases, food-safety procedures, forensic sciences and other applications.
|

