| Jan 30, 2013 |
Researchers unveil first artificial enzyme created by evolution in a test tube
|
|
(Nanowerk News) There’s a wobbly new biochemical structure in Burckhard Seelig’s lab at the University of Minnesota that may resemble what enzymes looked like billions of years ago, when life on earth began to evolve – long before they became ingredients for new and improved products, from detergents to foods and fuels.
|
|
Seelig created the fledgling enzyme by using directed evolution in the laboratory. Working with colleague Gianluigi Veglia, graduate student Fa-An Chao, and other team members, he subsequently determined its structure, which made its debut December 9 as an advance online publication in Nature Chemical Biology ("Structure and dynamics of a primordial catalytic fold generated by in vitro evolution").
|
|
Lab tests show that the enzyme (a type of RNA ligase, which connects two RNA molecules) functions like natural enzymes although its structure looks very different and it is flexible rather than rigid. Seelig speculates the new protein resembles primordial enzymes, before their current structures evolved.
|
|
Seelig and Veglia are professors in the College of Biological Sciences, where Fa-An Chao is a graduate student. Both faculty members have appointments in the Department of Biochemistry, Molecular Biology and Biophysics and Seelig is member of the BioTechnology Institute. Veglia also has an appointment in the Department of Chemistry in the university's College of Science and Engineering.
|
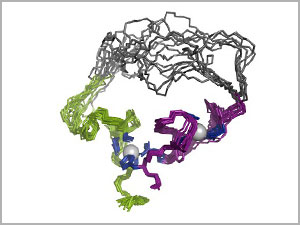 |
| 3-D structure of the evolved enzyme (an RNA ligase), using 10 overlaid snapshots. In the top region, the overlays show the range of bending and folding flexibility in the amino acid chain that forms the molecule. The two gray balls are zinc ions.
|
|
While a handful of groups worldwide are developing artificial enzymes, they use rational design to construct the proteins on computers. Instead, the Seelig lab employs directed evolution. "To my knowledge, our enzyme is the only entirely artificial enzyme created in a test tube by simply following the principles of natural selection and evolution," he says.
|
|
Rational enzyme design relies on preconceived notions of what a new enzyme should look like and how it should function. In contrast, directed evolution involves producing a large quantity of candidate proteins and screening several generations to produce one with the desired function. With this approach, the outcome isn’t limited by current knowledge of enzyme structure.
|
|
"Just as in nature, only the fittest survive after each successive generation," Seelig explains. The process continues until it produces an enzyme that efficiently catalyzes a desired biochemical reaction. In this case, the new enzyme joins two pieces of RNA together.
|
|
"It’s kind of like giving typewriters to monkeys," he says. "One monkey and one typewriter won’t produce anything clever. But if you have enough monkeys and typewriters, eventually one of them will write ’to be or not to be’." The lottery provides another analogy. "If you buy more tickets, you’re more likely to win," Seelig says.
|
|
Like all proteins, the new RNA ligase enzyme is a chain of amino acids folded into a 3D structure, but the resemblance stops there. Natural enzymes, like all proteins, are made from alpha helices and beta strands. Seelig’s artificial enzyme lacks those structures. Instead, it forms around two metal ions and is not rigid. "Compared to enzymes we know from nature, the artificial enzyme has a rather unusual structure and dynamics," Seelig says.
|
|
For decades, naturally occurring enzymes have been tweaked by industry to make industrial processes and products more effective. The ability to create enzymes from scratch using a natural process opens the door to a vast array of new products that provide business opportunities and improve quality of life without harmful environmental effects.
|
|
Going forward, Seelig plans to create enzymes with useful applications while he continues to explore the underlying basic science of enzyme structure and function, aiming to learn more about the origin of enzymes and how proteins evolve.
|
|
"Enzymes have always fascinated me," he says. "It’s rewarding to do work that has practical applications yet provides the opportunity to better understand how life on earth evolved."
|

