| Oct 23, 2013 |
Researchers get a detailed look at a DNA repair protein in action
|
|
(Nanowerk News) Errors in the human genetic code that arise from mismatched nucleotide base pairs in the DNA double helix can lead to cancer and other disorders. In microbes, such errors provide the basis for adaption to environmental stress. As one of the first responders to these genetic errors, a small protein called MutS – for “Mutator S” – controls the integrity of genomes across a wide range of organisms, from microbes to humans. Understanding the repair process holds importance for an equally impressive range of applications, including synthetic biology, microbial adaption and pathogenesis.
|
|
A new and detailed look at the role of MutS in DNA’s mismatch repair (MMR) system has been provided by a team of researchers with the U.S Department of Energy (DOE)’s Lawrence Berkeley National Laboratory (Berkeley Lab) and the Scripps Research Institute with their invention of a new technique for studying DNA. This breakthrough, which involves hybrid nanomaterials and small angle X-ray scattering (SAXS) technology, has been used to solve a major problem involving genome integrity and the biological detection of mismatched DNA.
|
|
Working at Berkeley Lab’s Advanced Light Source, the researchers used gold nanocrystal labels on DNA to create hybrid nanomaterials that are optimized for SAXS observation. The combination of gold-nanolabels and SAXS allowed the research team to follow DNA conformational changes brought on by MutS during the process of DNA mismatch error detection and response. They then showed that this hybrid nanolabel technique can also be used to examine short or long pieces of DNA in solutions that are comparable to cellular environments.
|
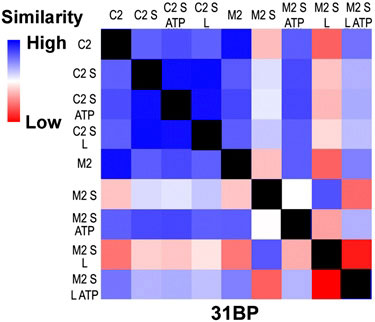 |
| This SAXS profile of a 31 base pair DNA with an error (M2) and no error (C2) in the presence of MutS (S), MutL (L), ATP and combinations. The profile is scored for pair-wise agreement and assigned a color ranging from high similarity (blue) to low similarity (red). The black squares are self-comparisons.
|
|
“Our technique of employing SAXS with gold nanolabels allows us to examine DNA processing by cooperative enzymes in which solution conditions, long distances, low concentrations, substoichiometric populations, and short time-scales are of importance,” says Greg Hura, a scientist with Berkeley Lab’s Physical Biosciences Division.
|
|
Hura is the lead author of a paper describing this research in the Proceedings of the National Academy of Sciences (PNAS). The article is titled “DNA conformations in mismatch repair probed in solution by X-ray scattering from gold nanocrystals”. The corresponding author is John Tainer, who holds joint appointments with Berkeley Lab’s Life Sciences Division and the Scripps Research Institute.
|
|
“It is a common belief that DNA is a passive component in protein interactions that involve DNA metabolism, but many proteins actually make use of DNA structural features, such as rigidity and conformation for important biological processes,” Tainer says. “The view of DNA as a passive element is at least in part due to a paucity of robust tools for examining dynamic DNA conformational states during multistep reactions.”
|
|
For this study, Tainer, Hura and their colleagues were able to capitalize on the high quality X-ray beams at ALS beamline 12.3.1, also known as SIBYLS, which stands for Structurally Integrated Biology for Life Sciences. Maintained by Berkeley Lab’s Life Sciences Division under the direction of Tainer, the SIBYLS experimental station is optimized for SAXS imaging, which provides global information on the conformations adopted by a population of macromolecules in almost any solution condition.
|
|
“Because X-rays scatter predominantly from electrons, the use of gold nanocrystals provides extremely high contrast relative to organic molecules critical for biology,” Tainer says. “This is important because we can follow specific biological molecules in complex reactions and along pathways to understand how their changes in shape and assembly control the biological outcomes.” |
|
The SAXS study at the SIBYLS beamline validated what has been dubbed the “beads-on-a-string” model of DNA repair, in which MutS proteins are the beads and DNA is the string. In solution, the MutS protein will bind to a mismatched DNA site by bending the DNA. ATP enzymes will come in to encircle and excise the error. The MutS then straightens out the bend and continues to proofread the DNA.
|
|
“This is the first time we used this technique to look at a protein-mediated process like DNA repair in solution with multiple partners,” Hura says. “We were able to determine some important details about MutS and the MMR system that should be valuable for drug design. We also now know what to look for in cancer-causing mutations of MutS. When we look at mutant versions of MutS we may be able to see that they do not bend the DNA or form the filament to the same extent as the normal version.”
|

