| Oct 16, 2014 |
A new self-assembly process allows the recognition of specific DNA sequences
|
|
(Nanowerk News) Transcription Factors (TFs) are specialized proteins that regulate gene expression by binding to specific DNA regulatory sequences, and thereby promote or inhibit the timely biosynthesis of particular proteins. Given their fundamental role in gene regulation it is not surprising that alterations in the activity of TFs are at the origin of many diseases, including cancer.
|
|
In many cases, their interaction with the DNA occurs as part of multimeric complexes (complexes involving more than one type of protein). Moreover, it is also known that the DNA recognition process by many TFs is coupled to the folding of their DNA-binding domains into defined structures.
|
|
As a result of their central biological role, there has been a great interest in the development of miniaturized synthetic models of TFs capable of reproducing the DNA binding properties of the natural proteins.
|
 |
|
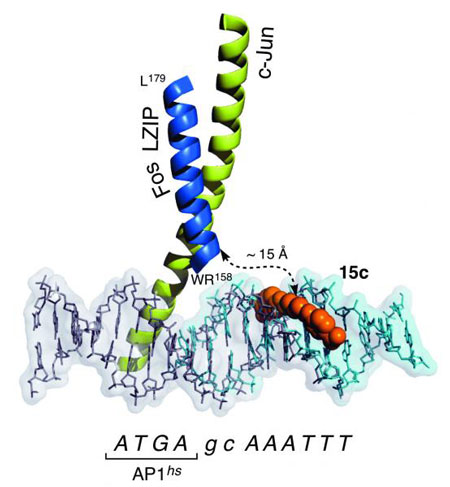
CiQUS researchers have introduced a new approach for achieving a highly selective, recognition of designed nine DNA base pairs ("Reversible Supramolecular Assembly at Specific DNA Sites: Nickel-Promoted Bivalent DNA Binding with Designed Peptide and Bipyridyl–Bis(benzamidine) Components"). The strategy involves the nickel-promoted assembly of a peptide derived from a transcription factor, and a small molecule equipped with a metal-binding unit that acts as heterodimerizing staple.
|
|
This system represents an infrequent case of self-assembly, as it involves four different components: a metal, a peptide, a small molecule and a nucleic acid. The multicomponent nature of the system, and the kinetic lability of the metal coordination, facilitates the disassembly of the supramolecular structure upon addition of external agents that sequester the nickel cation. Overall, the researchers have devised a programmed supramolecular system with emergent properties that reproduces some key characteristics of naturally occurring DNA binding proteins, such as bivalence, selectivity, responsiveness to external agents and reversibility.
|
|
This work has been developed by CIQUS the PhD students Jesús Mosquera and Mateo I. Sánchez, under the supervision of Prof. M. Eugenio Vázquez and José Luis Mascareñas, from the MetBioCat research group.
|

