| Mar 23, 2021 |
Mystery protein helps COVID-19 avoid immunity
|
|
(Nanowerk News) As COVID-19 rapidly spread around the globe in 2020, researchers sought explanations for the disease’s peculiar virulence. A team of HIV researchers, cellular biologists, and biophysicists banded together to determine the atomic structure of a certain protein thought to help SARS-CoV-2 (the coronavirus that causes COVID-19) evade and dampen the response from human immune cells.
|
|
In x-ray crystallography studies at the ALS, the researchers solved the structure of this protein, ORF8, and discovered two regions never before seen in other coronaviruses. These regions allow ORF8 to form large complexes, which may enable the virus to evade or suppress an immune response.
|
|
The findings have been published in PNAS ("Structure of SARS-CoV-2 ORF8, a rapidly evolving coronavirus protein implicated in immune evasion").
|
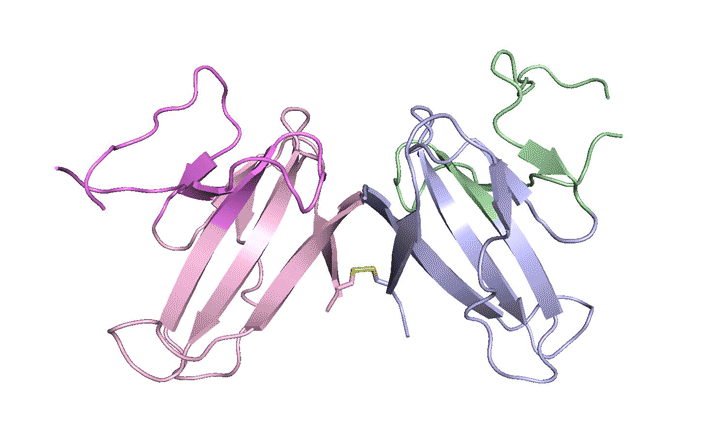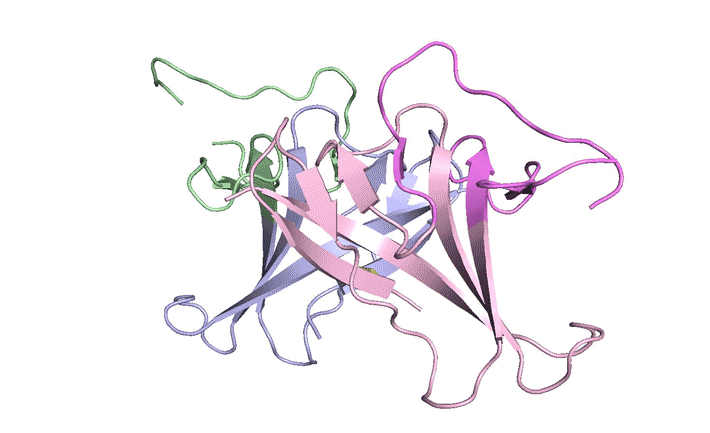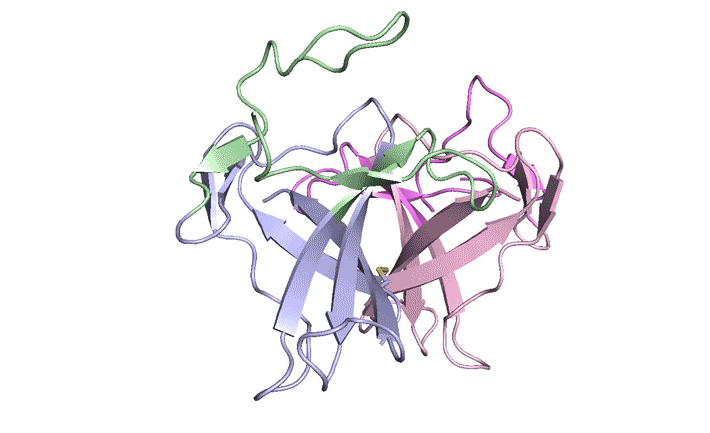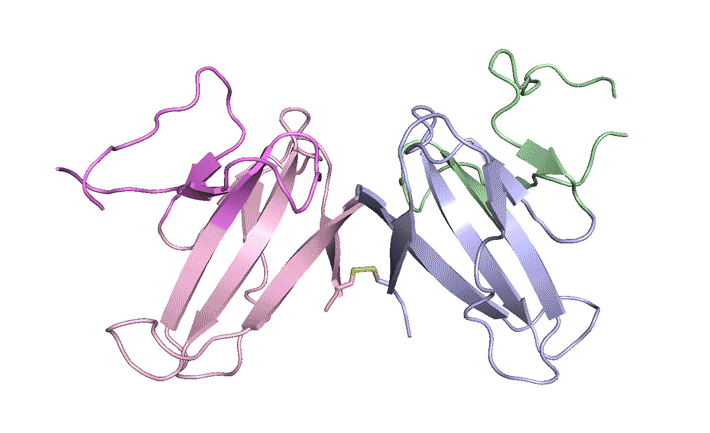 |
| A ribbon diagram of the ORF8 structure. This protein is composed of two units with identical amino acid sequences that are bound together by specific protein–protein interactions and a disulfide bridge. (Image: Berkeley Lab)
|
|
The work lays the groundwork for new antiviral treatments tailored to SARS-CoV-2 and enables further investigations into how the newly emerged virus ravages the human body.
|
A significant recombination mutation
|
|
Coronaviruses mutate differently from viruses like influenza or HIV, which quickly accumulate many little changes through a process called hypermutation. In coronaviruses, big chunks of nucleic acids sometimes move around through recombination. When this happens, big, new regions of proteins can appear.
|
|
Genetic analyses conducted very early in the pandemic revealed that SARS-CoV-2 evolved from a coronavirus that infects bats, and that a significant recombination mutation occurred in the area of the genome that codes for a protein, called ORF7, found in many coronaviruses.
|
|
The new form of ORF7, named ORF8, quickly gained the attention of virologists and epidemiologists because significant genetic divergence events like the one seen for ORF8 are often the cause of a new strain’s virulence.
|
|
For many proteins, the process of building a structural map is helped along by comparing the molecule’s structure to other proteins with similar amino acid sequences that have already been solved, allowing scientists to make informed guesses about how the protein folds into the 3D shape that determines its function. But ORF8’s amino acid sequence is so unlike any other protein that scientists had no reference for its overall shape, and they had to start from scratch.
|
Two unique regions
|
|
Using x-ray crystallography at ALS Beamline 5.0.2, part of the Berkeley Center for Structural Biology (BCSB), the researchers built an atomic model of ORF8, a process that required hundreds of crystals with multiple versions of the protein and thousands of diffraction images analyzed by special computer algorithms.
|
|
The results revealed two unique regions: one that is only present in SARS-CoV-2 and its immediate bat ancestor, and one that is absent from any other coronavirus. Essentially, the mutation caused the protein to double in size, in a way not related to any known fold. The regions stabilize the protein—which is a secreted protein, not bound to the membrane like the virus’s characteristic spike proteins—and create new intermolecular interfaces.
|
|
The group, and others in the research community, believe these interfaces are involved in reactions that somehow make SARS-CoV-2 more pathogenic than the strains from which it evolved.
|
|
By characterizing a completely new viral protein structure, these researchers have laid the groundwork for further studies into ORF8 interactions with antibodies and how therapies can target those interactions—crucial studies that are already underway.
|

