| Nov 01, 2012 |
Cellular landscaping: Predicting how, and how fast, cells will change
|
|
(Nanowerk News) A research team at the National Institute of Standards and Technology (NIST) has developed a model for making quantifiable predictions of how a group of cells will react and change in response to a given environment or stimulus—and how quickly ("Predicting rates of cell state change caused by stochastic fluctuations using a data-driven landscape model"). The NIST model, in principle, makes it possible to assign reliable numbers to the complex evolution of a population of cells, a critical capability for efficient biomanufacturing as well as for the safety of stem cell-based therapies, among other applications.
|
|
The behavior and fate of cells are only partially determined by their DNA. A living cell reacts to both its internal and external environment—the concentration of a particular protein inside itself or the chemistry of its surroundings, for example—and those reactions are inherently probabilistic. You can't predict the future of any given cell with certainty.
|
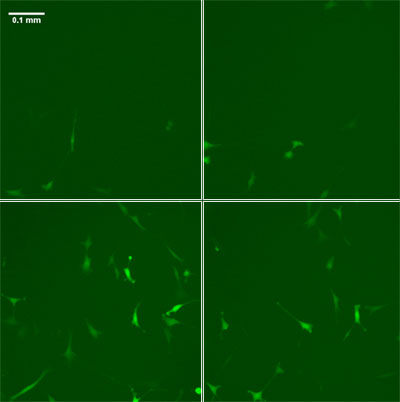 |
| Time lapse fluorescence images of a cell culture (clockwise from top left, start - 14 hours - 28 hours - 42 hours) reveal how the expression of a particular gene in the culture varies not only from cell to cell, but with time.
|
|
This inherent uncertainty has consequences, according to NIST biochemist Anne Plant. "In the stem cell area in particular, there's a real safety and effectiveness issue because it's very hard to get 100 percent terminal differentiation of stem cells in a culture," she says. This could be problematic, because a therapist wishing to produce, say, heart muscle cells for a patient, might not want to introduce the wild card of undifferentiated stem cells. "Or effectiveness may be dependent on a mixture of cells at different stages of differentiation. One of the things that is impossible to predict at the moment is: if you waited longer, would the number of differentiated versus nondifferentiated cells change? Or if you were to just separate out the differentiated cells, does that really remove all the nondifferentiated cells? Or could some of them revert back?" says Plant.
|
|
The NIST experiments did not use stem cells, but rather fibroblasts, a common model cell for experiments. The team also used a standard tracking technique, modifying a gene of interest—in this case, one that codes for a protein involved in building the extracellular support matrix in tissues—by adding a snippet that codes for a small fluorescent molecule. The more a given cell activates or expresses the gene, the brighter it glows under appropriate light. The team then monitored the cell culture under a microscope, taking an image every 15 minutes for over 40 hours to record the fluctuations in cell behavior, the cells waxing and waning in the degree to which they express the fluorescent gene.
|
|
Custom software developed at NIST was used to analyze each image. Both time-lapse data from individual cells and time-independent data from the entire population of cells went into a statistical model. The resulting graph of peaks and valleys, called a landscape, says Plant, "mathematically describes the range of possible cell responses and how likely it is for cells to exhibit these responses." In addition, she says, the time analysis provides kinetic information: how much will a cell likely fluctuate between states, and how quickly?
|
|
The combination makes it possible to predict the time it will take for a given percentage of cells to change their characteristics. For biomanufacturing, it means a finer control over cell-based processes. If applied to stem cells, the technique could be useful in predicting how quickly the cells differentiate and the probability of having undifferentiated cells present at any point in time.
|

