| Apr 12, 2013 |
New atomic force microscopy technique to decode secrets of our gut
|
|
(Nanowerk News) A new technique based on atomic force microscopy was developed at the Institute of Food Research to help ‘read’ information encoded in the gut lining.
|
|
The lining of our gut is an important barrier between the outside world and our bodies. Laid out, the gut lining would cover the area of a football pitch. It must let nutrients from our foods through, but prevent invasion by disease-causing bacteria, at the same time hosting the trillions of beneficial bacteria needed for proper digestion and immune function.
|
|
mucinAt the forefront of the defensive system is a layer of mucus that lines the entire gut surface. In the large bowel, the mucus layer is an organised structure, with an inner layer that blocks bacteria, and an outer layer where beneficial bacteria (commensals) can flourish. The mucus layer is made up of large proteins called mucins with characteristic sugary, or glycosylated, chains.
|
|
Dr Nathalie Juge and colleagues from the Institute of Food Research, which is strategically funded by BBSRC, is leading studies aiming to understand the role of mucus in maintaining a healthy gut and how mucins interact with bacteria in our guts.
|
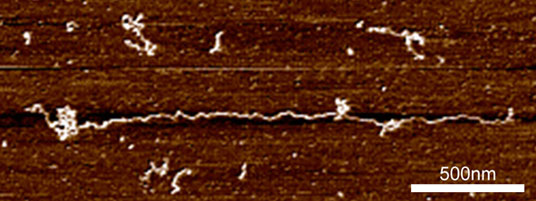 |
| An extended mucin chain on a mica surface.
|
|
At the molecular level, mucins exhibit clusters of glycosylation that give the proteins a ‘bottle-brush’ appearance. The size of these molecular sugar chains varies with location and age of the tissue, and abnormalities in mucins are seen in inflammatory bowel diseases such as ulcerative colitis, Crohn’s disease and colon cancer. A myriad of different conformations that the sugar chains can take up means that mucins are incredibly diverse. The differences are believed to help commensal bacteria by providing specific binding sites, as well as necessary nourishment.
|
|
Pathogenic bacteria have also evolved mechanisms to bind to mucins, as part of their arsenal to overcome our defences. We, however, are a long way behind these bacteria in understanding the coded information contained in these complex sugar chains. Understanding this ‘glycocode’ would give us a new way to look at the crucial interactions between good and bad microorganisms in our guts, and may also provide new insights into gut diseases.
|
|
These interactions happen at the molecular level, so for us to understand them we need tools that work at this level. IFR has a history of pioneering use of one such tool, atomic force microscopy (AFM).
|
|
AFM combines very high-resolution imaging with an ability to probe the forces between molecules. AFM works by running a very fine stylus (tip) mounted on the end of a flexible cantilever over the surface of a molecule, much like a blind person reading braille. A laser is bounced off the cantilever, amplifying the signal so that AFM can detect distances down to a millionth the width of a sheet of paper.
|
|
Patrick Gunning and Andrew Kirby, from IFR’s AFM group, adapted this technique by attaching sugar-binding molecules called lectins to the AFM cantilever, via a flexible linker. They used this to probe mucins bound to a surface. The findings of this collaborative work were published in the FASEB Journal ("Mining the 'glycocode'—exploring the spatial distribution of glycans in gastrointestinal mucin using force spectroscopy").
|
|
“It’s a bit like fishing” said Dr Gunning. “The mucin molecules are immersed in saline, and float like a sea of kelp. We use lectins as the bait. We drop the line down until we hit the bottom, and then lift it back up. If the lectin finds a target sugar molecule on the mucin, it snags.”
|
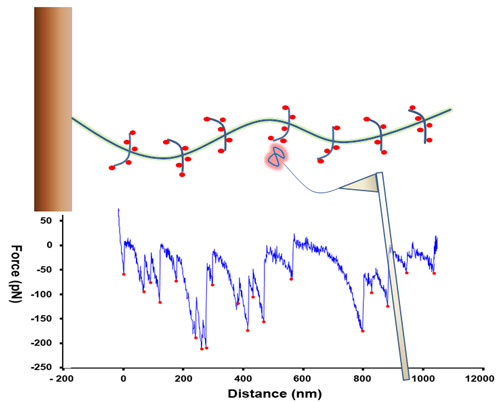 |
| Schematic diagram showing a mucin molecule being probed by a lectin bound to an AFM cantilever. When the lectin ‘snags’ on sugar, the cantilever registers this. The spatial arrangement of these signals can be used to fingerprint the mucin.
|
|
Measuring the distance between the snags gives a picture of what the overall mucin molecule looks like. By repeating this thousands of times, and then working with IFR’s biomathematicians, it was possible to produce a ‘fingerprint’ that characterise different mucins, which means that we could differentiate between mucins derived from different parts of the gut.
|
|
The researchers now want to look at the mucins derived from diseased tissues, to further investigate differences in the glycocode. They would also like to understand how bacteria read and possibly manipulate the glycocode. In the lab, the researchers used specific enzymes targeting certain sugars on the mucin molecules. This affected the mucin’s molecular structure, which in turn altered its spatial distribution. Certain bacteria in our guts also secrete these enzymes, as a means of rewriting the glycocode for their own benefit. With this new technique in hand, the researchers will continue their efforts to unravel the secrets inside our own bodies.
|


