| Sep 12, 2013 |
NIH award combines plasmonic light focusing with nanopore technology for DNA sequencing
|
|
(Nanowerk News) An international team of researchers from the University of Illinois at Urbana-Champaign and Delft University of Technology (TU Delft) in the Netherlands is advancing a novel form of nanopore technology for DNA sequencing as a result of a $2.47 million grant awarded by the National Institutes of Health (NIH).
Nanopore-based DNA sequencing concepts generally entail one of the DNA strands passing through the nanopore sensor, where the individual nucleotides are distinguished from each other.
|
|
The grant is part of the $17 million awarded to eight research teams through the NIH National Human Genome Research Institute (NHGRI)'s Advanced DNA Sequencing Technology program, which was launched in 2004.
|
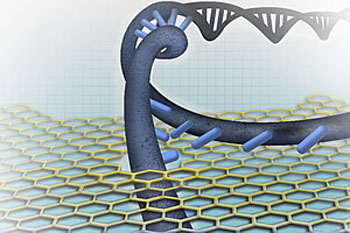 |
| Nanopore-based DNA sequencing concepts generally entail one of the DNA strands passing through the nanopore sensor, where the individual nucleotides are distinguished from each other.
|
|
"Nanopore technology shows great promise, but it is still a new area of science. We have much to learn about how nanopores can work effectively as a DNA sequencing technology, which is why five of the program's eight grants are exploring this approach," said Jeffery A. Schloss, program director for NHGRI's Advanced DNA Sequencing Technology program and director of the Division of Genome Sciences.
|
|
Nanopore-based DNA sequencing involves threading single DNA strands through tiny pores. Individual base pairs—the chemical letters of DNA—are then read one at a time as they pass through the nanopore.
|
|
According to Schloss, this technology offers many potential advantages over current DNA sequencing methods, including real-time sequencing of single DNA molecules at low cost and the ability for the same molecule to be reassessed over and over again.
|
|
“This research project aims to combine the unique and powerful capabilities of two exciting, rapidly evolving fields—plasmonics and nanopores—for the analysis of single DNA molecules,” explained Aleksei Aksimentiev, an associate professor of physics at Illinois and principle investigator for the project. “More specifically, recent advances in nanoplasmonics will be utilized to enable label-free , single-molecule trapping and sequencing of DNA using nanopores.
|
|
“To our knowledge we are among the first groups to explore this powerful combination of nanopores and plasmonics,” Aksimentiev added. “We will explore this using the combination of molecular dynamics simulations here at Illinois, with the experiments supervised by Magnus Jonsson and Cees Dekker, our co-PIs at TU Delft.”
|
|
“We use 'nanopores' as sensors,” stated Dekker, who chairs the Department of Bionanoscience at TU Delft. “These nanopores are less than a millionth of an inch in size, through which we let a biomolecule such as DNA flow. Upon feeding the molecule through the hole we aim to read off information about it. Our ultimate goal is to read off the sequence of a single DNA molecule in this way.”
|
|
“The novel ingredient of our new sensor is a gold nanostructure that acts as an optical antenna and allows us to focus light to a highly intense nanoscale region right at the nanopore,” explained Jonsson, who is the third co-PI of the project and a postdoc in Dekker’s lab at TU Delft. “DNA fragments moving through the nanopore will be exposed to the strong optical fields in this ‘plasmonic hot spot’, offering unique means for both controlling the motion of the DNA and for optical readout of the DNA sequence.”
|
|
This novel approach to advancing DNA through the nanopore simultaneously enables DNA sequence detection through surface-enhanced Raman spectroscopy. Because locally confined plasmonic fields enhance Raman scattering many orders of magnitude and because of the direct relationship of Raman spectra to the underlying molecular structure, sequence detection will be possible directly, without any labeling.
|
|
According to the NIH, costs of DNA sequencing have greatly declined since 2003, when the genome sequencing performed under the Human Genome Project was completed at a cost of approximately $1 billion. Only a year later, in 2004, sequencing a human genome cost an estimated $10-50 million, thanks to improvements in technologies and tools. By 2009, NHGRI met its goal of producing high-quality human genome sequences at a 100-fold reduction in price, or $100,000. While achieving another 100-fold drop in price has been difficult, sequencing a person's genome today costs about $5,000 to $6,000.
|
|
NHGRI is one of the 27 institutes and centers at the National Institutes of Health, the nation's medical research agency. The NHGRI Extramural Research Program supports grants for research and training and career development at sites nationwide. Additional information about NHGRI can be found at www.genome.gov.
|

