| Posted: Sep 18, 2017 | |
A DNA-based rewritable memory device |
|
| (Nanowerk Spotlight) DNA is well known as the genetic material, but has also been used as a building block for the construction of nanoscale structures and periodic arrays. One other application is the storage of information in DNA, with tech giants such as Microsoft joining in the bandwagon to develop it further. | |
| In short, the information to be stored is coded in "DNA language", i.e. a specific sequence of the four nucleotides (or bases) of the molecule: adenine, thymine, cytosine and guanine. DNA strands with these specific sequences are then synthesized and stored as libraries. The stored information is retrieved by sequencing the pools of DNA strands followed by decoding the message. | |
| Such an information storage method has many advantages including longevity (DNA has a half-life of ∼500 years) and ability for highly dense storage (∼1 million terabytes/mm3), and is more suited for archival storage of data. | |
| However, reading only portions of the stored message is hard since entire libraries of DNA strands have to be sequenced. Also, sequencing DNA is an elaborate and (for now) costly method. | |
| In our recent paper in Nucleic Acids Research ("Addressable configurations of DNA nanostructures for rewritable memory"), we use shape-changing DNA nanostructures for short term storage of data, where the information is "written" in different conformations of the nanostructures. The stored data can be easily read-out using gel electrophoresis, eliminating any multi-step or costly methods. | |
| Moreover, the written information can be erased and rewritten, with our strategy also providing a write-protection function (to prevent erasing of specific bits). | |
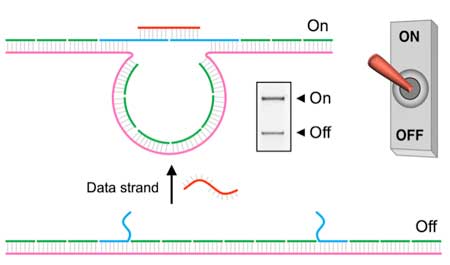 |
|
| The DNA nanoswitch used in our study can be triggered using specific DNA strands. The "off" state of the device is a linear duplex formed by a long single stranded scaffold strand (∼7 kilobases) hybridized to short complementary backbone oligonucleotides. Two of the backbone oligonucleotides can be modified to contain single-stranded extensions that are partly complementary to another DNA strand (this serves as the input data strand). Binding of the data strand to the nanoswitch creates a looped "on" state that can be easily identified on a gel (see figure above). | |
| We expanded this system to serve as a 5-bit memory device by incorporating six address site oligonucleotides on the scaffold strand separated by about 600 base pairs (or 200 nm). | |
| The first address site is common to all data strands (i.e. complementary to one-half of all the data strands) and each of the other address sites are specific to five different data strands. Combinations of the five data strands can be used to create different looped states (of different loop sizes), each of which acts as a "bit" on the gel lane (see figure below). | |
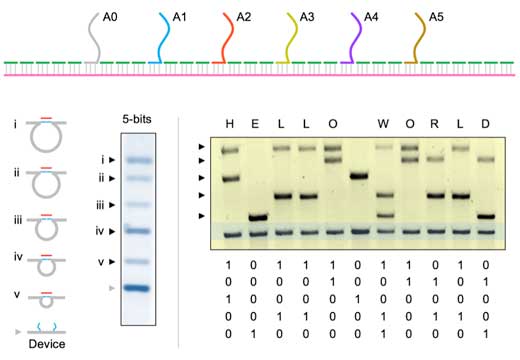 |
|
| We encoded the words Hello World using this system, erased it using toehold-mediated strand displacement (where the data strand is removed using a fully complementary DNA strand), and rewrote Good Bye on the same system. | |
| Our strategy provides write-protection by choosing data strands without toe-holds so that specific bits can be prevented from being erased in sequential erasing or rewriting steps. | |
| Moreover, the device is stable and functional even after drying and can thus be potentially used for labeling with 5-bit codes, or expanded to 8-bits or more (several bytes) by placement of more address sites. The ease of use and readout also makes this system feasible for data encryption. | |
| Since the system is based on identifying specific DNA strands, it can be used in biosensing for multiplexed detection of nucleic acid sequences with unique outputs for specific biomarkers. | |
|
By Arun Richard Chandrasekaran
|
|
|
Become a Spotlight guest author! Join our large and growing group of guest contributors. Have you just published a scientific paper or have other exciting developments to share with the nanotechnology community? Here is how to publish on nanowerk.com. |
|
