| Aug 18, 2021 | |
Intracellular activation of RNA interference upon DNA-driven organization of quantum dot assemblies |
|
| (Nanowerk Spotlight) In addition to the plethora of functions such as storage of genetic information and regulation of its expression, DNA and RNA are also highly programmable biomaterials. | |
| Stemming from their predictable Watson-Crick base pairing (G=C and A-T), DNA can be utilized to design short complementary sequences to be used as the linkers which bring together and organize other biological and inorganic materials. | |
| Quantum dots (QDs) are one such inorganic candidate. The unique optical properties of these semiconductor nanoparticles have been previously utilized in a number of applications, including for imaging and biosensing when brought together in bulk. | |
| In a recent collaboration between researchers at the University of North Carolina at Charlotte, North Carolina State University, Columbia University, and Frederick National Laboratory for Cancer Research, an interdisciplinary team utilized DNA for the precise assembly of QDs into larger three-dimensional scaffolds. | |
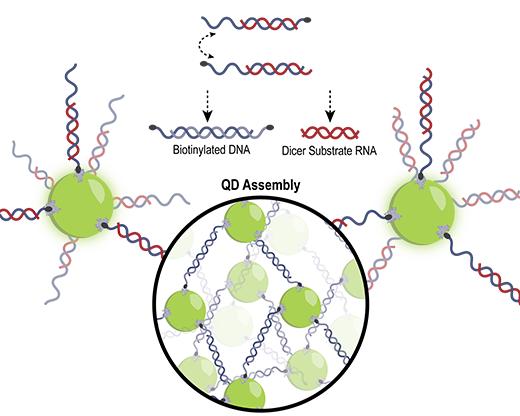
| To achieve this, the DNA sequences were functionalized with biotin, which was then used to link together streptavidin-decorated QDs emitting at 545 nm. | |
| The team reported their findings in ACS Applied Materials & Interfaces ("Controlled Organization of Inorganic Materials Using Biological Molecules for Activating Therapeutic Functionalities"). | |
| “We developed a simple and user-friendly combinatorial approach for intracellular material organization with nanoscale precision,” Professor Kirill Afonin from the Department of Chemistry, University of North Carolina at Charlotte, tells Nanowerk. “We also implemented some therapeutically relevant modules that can be activated in response to this organization.” | |
 |
|
| DNA/RNA hybrid duplexes drive the assembly of QDs and release of therapeutic RNAs. (Image courtesy of the researchers) | |
| Three different approaches for QD assembly were assessed: adding a double-biotinylated DNA duplex to QDs; adding two QDs each with complementary single-stranded DNAs; and finally, adding two QDs each with complementary DNA/RNA hybrid duplexes. | |
| While all approaches resulted in the assembly of QDs, they all exhibited differences in the kinetics of their assembly. | |
| The assembled QD formations were assessed by researchers at three different institutions using electrophoretic mobility shift assays, transmission electron microscopy (TEM), and small-angle X-ray scattering (SAXS) which informed the one-, two-, and three-dimensional properties of the assemblies, respectively. Interparticle spacing between QDs was consistent across TEM and SAXS, and highly ordered compared to unassembled QDs. | |
| To investigate the assembly in cells, researchers transfected two types of QDs with complementary DNA linkers into a human breast cancer cell line and were able to observe the colocalization of both emissions using confocal microscopy. | |
| Furthermore, the DNA/RNA hybrid approach was utilized in order to introduce therapeutic RNAs into the cellular environment upon QD assembly. Driven by a toehold-mediated isothermal reassociation of the DNA sequences, the complementary RNAs, which were kicked off, were designed to silence green fluorescent protein (GFP) via RNA interference. Upon assembly in a cell line expressing GFP, the QD assemblies were additionally confirmed through the activation of the therapeutic response. | |
| Importantly, the QD assemblies also showed low cytotoxicity and immune stimulation when introduced into these human cell lines. | |
| Source: Provided by the University of North Carolina Charlotte as a Nanowerk exclusive | |
|
Become a Spotlight guest author! Join our large and growing group of guest contributors. Have you just published a scientific paper or have other exciting developments to share with the nanotechnology community? Here is how to publish on nanowerk.com. |
|
