| Feb 28, 2022 |
An expanded molecular alphabet for DNA data storage
(Nanowerk News) DNA data storage systems have the potential to hold orders of magnitude more information than existing systems of comparable size. Compared to existing data storage technologies, it is potentially less expensive, far more physically compact, more energy efficient, and longer lasting – DNA survives for hundreds of years and doesn't require maintenance. Files stored in DNA also can be very easily copied for negligible cost.
|
|
DNA’s storage density is staggering. Consider this: humanity will generate an estimated 33 zettabytes by 2025 – that is 3.3 followed by 22 zeroes. All that information would fit into a ping pong ball, with room to spare. The U.S. Library of Congress has about 74 terabytes, or 74 million million bytes, of information – 6,000 such libraries would fit in a DNA archive the size of a poppy seed.
|
|
Information stored in DNA can be copied in a massively parallel manner and selectively retrieved via polymerase chain reaction (PCR). However, existing DNA storage systems suffer from high latency caused by the inherently sequential writing process. Despite recent progress, a typical cycle time of solid-phase DNA synthesis is on the order of minutes, which limits the practical applications of this molecular storage platform.
|
|
To overcome these challenges, new synthesis methods and information encoding approaches are required to accelerate the speed of writing large-volume data sets.
|
|
Expanding the alphabet of a DNA storage media by including chemically modified DNA nucleotides can both increase the storage density and the writing speed because more than two bits are recorded during each synthesis cycle.
|
|
Advancing the research in this regard, scientists have now reported an expanded molecular alphabet for DNA data storage comprising four natural and seven chemically modified nucleotides that are readily detected and distinguished using nanopore sequencers.
|
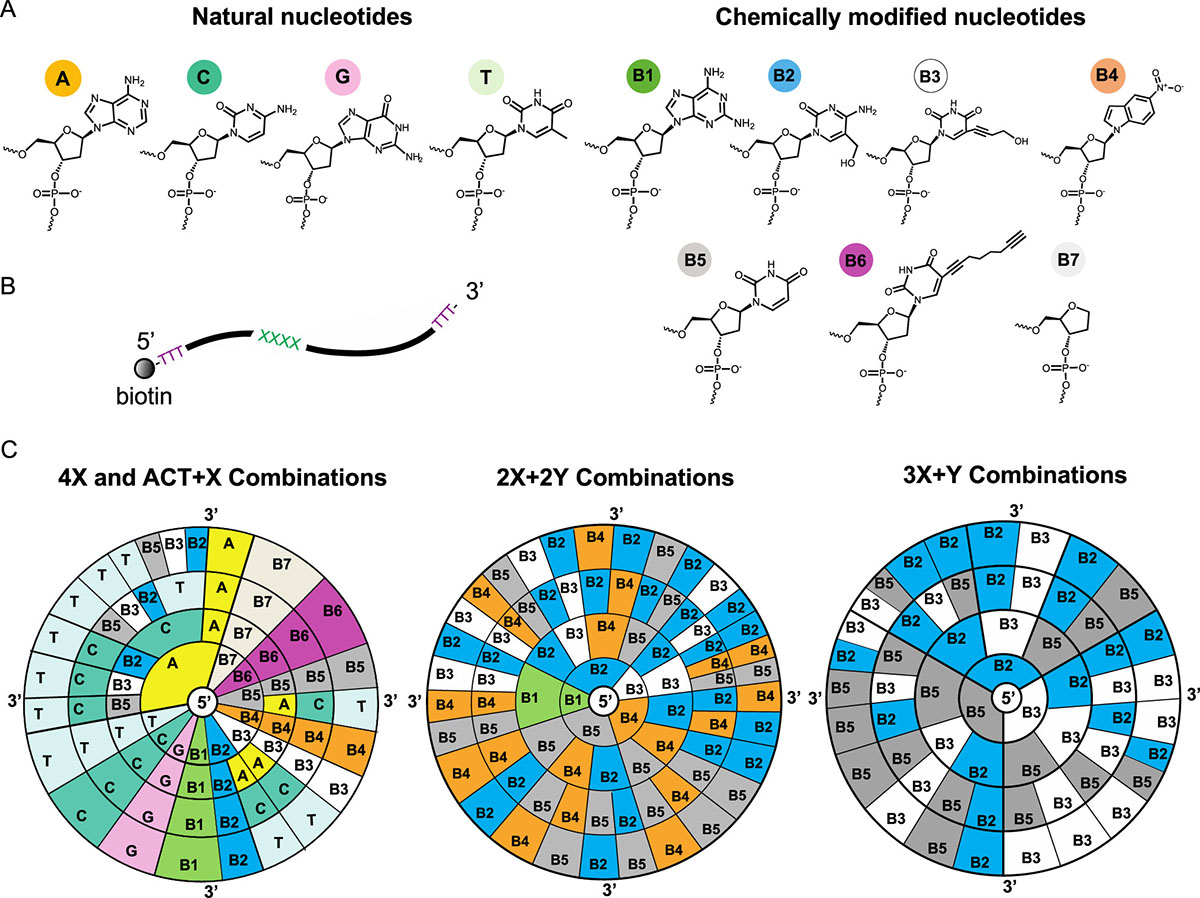 |
| DNA data storage using natural and chemically modified nucleotides. (A) Chemical structures of natural DNA nucleotides (A, C, G, T) and the selected chemically modified nucleotides employed in our study (B1−B7). (B) Schematic of the ssDNA oligo used in MspA nanopore experiments. The length of the oligos is 40 nucleotides (nts), with biotin attached at the 5′ terminus. Homo- or heterotetrameric sequences are located at positions 13−16, flanked by two polyT regions of length 12 nt and 24 nt on the 5′ and 3′ ends, respectively. (C) Sequence space for DNA homotetramers or heterotetramers used in MspA nanopore experiments. The notation aX + bY, where a and b take values in {2, 3, 4} so that a + b = 4, indicates that “a” symbols of the same kind are combined with “b” symbols of another kind and arranged in an arbitrary linear order. In total, 77 distinct tetrameric sequences were synthesized and tested experimentally. (Left) Circular diagram showing all 11 homotetramers and 12 tetrameric sequences of the form ACT + X, where X is a chemically modified nucleotide from the set {B2, B3, B5}. (Middle) Circular diagram showing all 30 tested combinations of tetrameric sequences with total composition 2X + 2Y using chemically modified monomers from the set {B1, B2, B3, B4, B5}, including sequence patterns XXYY, XYYX, and XYXY. (Right) Circular diagram showing the remaining 24 combinations of tetrameric sequences with total composition 3X + Y using the set {B2, B3, B5}. Five chemically modified nucleotides form stable base pairs with natural nucleotides via hydrogen bonds (B2−G, B3−A, B5−A, B6−A, B6−C), based on the results from molecular dynamic (MD) simulations. (Reprinted with permission by American Chemical Society) (click on image to enlarge)
|
|
The findings are published in Nano Letters ("Expanding the Molecular Alphabet of DNA-Based Data Storage Systems with Neural Network Nanopore Readout Processing").
|
|
The authors'results show that Mycobacterium smegmatis porin A (MspA) nanopores, which are widely used for ssDNA sensing and single molecule chemistry studies, can accurately discriminate 77 combinations and orderings of chemically diverse monomers within homo- and heterotetrameric sequences.
|
|
They further demonstrate that highly accurate classification (exceeding 60% on average) of combinatorial patterns of natural and chemically modified nucleotides is possible using deep learning architectures.
|
|
According to the scientists, the extended molecular alphabet has the potential to offer a nearly 2-fold increase in storage density and potentially the same order of reduction in recording latency, thereby providing a promising path forward for the development of new molecular recorders.
|

 By
Michael
Berger
– Michael is author of three books by the Royal Society of Chemistry:
Nano-Society: Pushing the Boundaries of Technology,
Nanotechnology: The Future is Tiny, and
Nanoengineering: The Skills and Tools Making Technology Invisible
Copyright ©
Nanowerk LLC
By
Michael
Berger
– Michael is author of three books by the Royal Society of Chemistry:
Nano-Society: Pushing the Boundaries of Technology,
Nanotechnology: The Future is Tiny, and
Nanoengineering: The Skills and Tools Making Technology Invisible
Copyright ©
Nanowerk LLC
