| Aug 01, 2012 |
Getting to the root - unearthing the plant-microbe quid pro quo
|
|
(Nanowerk News) While the flower may attract the bee and the admiring eye of the passerby, it is the unseen complex network of life below ground where the action is. The microbial community or microbiome that inhabits the rhizosphere and endosphere —the niches immediately surrounding and inside a plant's root—facilitates the shuttling of nutrients and information into and out of the roots within the soil matrix. This commerce, by enabling the plant to adapt to its immediate environment, contributes to plant health and growth, defense against pests, management of carbon and other elements for the enrichment of the soil, and even informs the plant's ability to sequester contaminants. These underground microbial activities have not received as much attention as the effort to characterize the role of the microbial populations inside and on the surfaces of humans, but this is now changing owing to a recent publication in the journal Nature ("Defining the core Arabidopsis thaliana root microbiome").
|
|
Led by researchers from the U.S. Department of Energy Joint Genome Institute (DOE JGI) and the University of North Carolina (UNC), Chapel Hill, the study published August 2, 2012 sheds light on the mechanisms driving the subterranean formation of this "plant microbiome" and how plants can influence the presence of the microbiota in the rhizosphere and vice-versa.
|
|
"Understanding the rules that guide formation of the root microbiome are likely to contribute significantly to the success of agriculture and our understanding of the carbon cycle," said senior author Jeff Dangl, Howard Hughes Medical Institute (HHMI) investigator and UNC's John N. Couch Professor of Biology. "Science has long been fascinated with the spectrum of relationships between plant and microbe that span from pathogenic to mutually beneficial. With our results we are adding new details to this complex landscape."
|
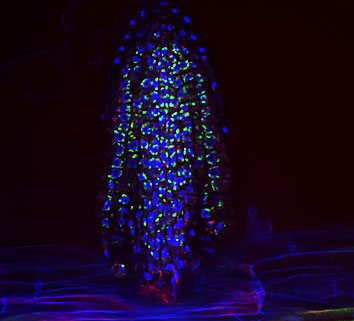 |
| An emerging lateral root covered in bacteria (shown in green).
|
|
The team, which also included researchers from University of Bremen, in Germany, Cornell University, and the University of Queensland in Australia, was able to pinpoint the identity of many of the microbial players so their metabolic roles could be inferred, as well as where and when they were pressed into service.
|
|
"Our study provides a deeper and more fine-tuned indication of how host plants and soil microbes shape the shared microbiome—we defined microbial taxa that are reproducibly attracted to plant roots, and which ones the plant allows to penetrate into the roots, presumably in exchange for nutritional or other benefits," Dangl said.
|
|
The team's approach combined the Dangl lab's robust experimental design with the power of the DOE JGI's deep genome sequencing and analysis of complex microbial communities. Co-author Susannah Tringe, head of DOE JGI's Metagenome Program, said the microbiome can be viewed as an extension of the plant's genome. "The rhizosphere is a dynamic microcosm of interdependence. We can't really know a plant genome's full functional capacity until we also understand the functional capacity and the drivers governing assembly of its associated microbiome. In the same way that microbes play critical roles in and around our own bodies, we are adopting this concept of host-associated metagenomics in plant genomics as well, as it will ultimately lead to predictive interventions that will increase plant health and productivity, disease resistance and carbon capture."
|
|
To get the lay of the under-land, the researchers employed a rigorous assessment of the endophytes—those microbes that exist within the plant-roots—as well as those microbes at the soil-root interface and in the surrounding soil. The findings, the team suggests, could facilitate development of a strategy of community-based plant probiotics. For instance, instead of spraying pesticides or fertilizer treatments, it would be possible to suggest to farmers which microbial species, and in which proportions, would best improve productivity in the crop plant of interest in a particular soil type.
|
|
The ambitious experiment targeted the microbial communities from over 600 Arabidopsis thaliana plants. This tiny, weedy, so-called "lab rat" of the plant kingdom was the first plant to have its genome sequenced, and grows quickly and easily in the lab. Plants were grown in two different soil types, then full plant root systems were removed from their pots and shaken to remove non-rhizosphere soil. Rhizosphere soil was then washed off and collected, and finally the roots themselves were frozen and ground up to access the microbes deep inside. The researchers then isolated DNA from these three compartments—soil, rhizosphere and endophytes—and looked at the 16S rRNA gene, a diagnostic tag that can help distinguish between microbial species. In a computational tour de force, the researchers sifted through massive amounts of sequence data from the soil, the root-soil interface and the root-cell compartment—over 1,200 samples constituting some two billion bases of high quality DNA code—to assemble the puzzle by developing a systematic model that enabled the team to quantify the contribution from each variable to the community composition.
|
|
The researchers proceeded on the assumption that plant roots, through their physical structure and the release of molecules, create a local environment that is chemically and microbiologically distinct from the soil in which they grow.
|
|
"We found that microbial endophyte communities are more dependent on the individual host plant 'genotype' than the rhizosphere communities," Dangl said. "Several microbes were consistently more common on and in roots than in soil, suggesting reproducible colonization and potential symbiosis. We also confirmed the expected result that all rhizospheres and endophytic compartments, regardless of soil type or genotype, assemble a microbial community distinct from soil. And our results suggest that a subset of the soil bacterial population is typically enriched in rhizosphere samples by biophysical and/or biochemical cues issued by the plant."
|
|
The team also found that some bacteria common in soil are depleted inside the root cells, suggesting that these microbes are either actively gated by the plant's immune system, or outcompeted by more successful root-cell colonizers.
|
|
"The microbial balancing act that we observed suggests a set of core ecological principles shaping the A. thaliana root microbiome," said Dangl. "We see an array of different microorganisms representing different phyla providing broad metabolic potential, while a certain subset is dependent on the particular genotype of plant and serves to individualize the metagenome."
|
|
"This opens a wide door through which we will define the microbiome's functional capacity, and the host genes that potentially contribute to microbial association phenotypes – genes of major agronomic interest." Tringe said. "The microbiota that inhabit this niche can both benefit and undermine plant health. Shifting this balance can influence crop success. For instance, some microbes may provide the plant with nutrients and hormones that can improve plant growth, suppress pests and increase resistance to stresses such as drought, while others may be stealthy pathogens."
|
|
Rhizosphere metagenomic efforts have been steadily gaining interest and support throughout the plant and microbial research communities. Jeff Bennetzen, professor at the University of Georgia, whose laboratory works on such related topics such as metagenomics for lignocellulosic biofuel production, concurred. "This work is the first step in a massive and exciting venture into the terra incognita of what genes are present in host genomes that determine the status of microbial populations near their roots."
|
|
The rhizosphere project grew out of recommendations from a 2008 study by the National Research Council of the National Academies entitled, "The Achievements of the National Plant Genome Initiative [NPGI] and New Horizons in Plant Biology." The publication noted that "genome sequence is the raw material for functional, evolutionary and translational tool development at the center of plant genome sciences," and that, "similar large-scale investment in plant-associated metagenomics is justified because of the diversity of plant-associated microbial communities and their impact on plant productivity…[f]or example, the communities of microorganisms associated with candidate perennial biofuels crops." The study also acknowledged that, "the unique role played by the Department of Energy's Joint Genome Institute (JGI) in the service of NPGI is critical."
|
|
The team is now engaged in an even larger-scale, long-term study supported by the HHMI, the Gordon and Betty Moore Foundation, the National Science Foundation and the DOE Office of Science to define the root-associated microbiomes of three important model plant species across a wide diversity of geographical locations and ecological conditions.
|

