| Nov 16, 2021 |
The Sorting Hat: An AI-powered image classifier for cell biologists
|
|
(Nanowerk News) Cell division is an important process that underlies biological growth and repair. Cell biologists track this process by observing chromosomes—structures made of DNA that contain the genetic material of an organism.
|
|
Advances in microscopy along with automation have allowed researchers to snap better images of chromosomes in a short time. However, their analysis is still largely done manually, which is often a tedious task. This is especially true for plants, which show a huge variety in chromosome sizes and numbers.
|
|
Now, in a recent study published in Chromosome Research ("Effectiveness of Create ML in microscopy image classifications: a simple and inexpensive deep learning pipeline for non-data scientists"), researchers from Japan have taken a different approach. Led by Associate Professor Kiyotaka Nagaki from Okayama University, Japan, they have used deep learning artificial intelligence (AI) to classify chromosomal images from several plant species.
|
|
While this in itself is nothing new, what is interesting is that the team has shown that it is possible for even non-experts to make use of AI readily.
|
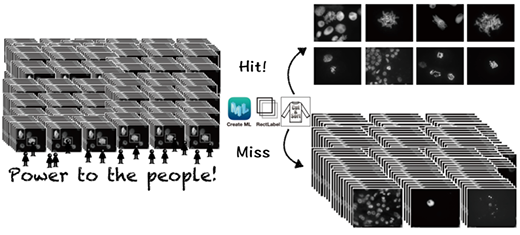 |
| Application of deep learning artificial intelligence (AI) for image classification requires a high level of expertise in machine learning. Now, researchers from Japan have created an AI image classifier that can be customized and readily used by non-experts. (Image: Kiyotaka Nagaki, Okayama University)
|
|
How was this made possible? Dr. Nagaki explains, “Classifying images using AI usually requires a high level of computer knowledge. What we did is build AI models on a McIntosh computer with the CreateML app suitable for our own image samples. Moreover, the AI can be trained to become an order-made image classifier for any variety of images that suits one’s purpose.”
|
|
The team used chromosomal images to train the deep learning models to detect images or parts of images where the cells are undergoing “mitosis,” a process where a single cell divides into two identical daughter cells. They estimated its detection accuracy with test images based on the number of cells the model correctly classified.
|
|
Next, the team put the models to test with images containing mitotic cells from plant species not used during the training. To their delight, the models correctly distinguished mitotic cells in these images. In addition, the technique also worked well for cells in tissue sections and a different process of cell division.
|
|
These results indicate that the deep learning pipeline developed by the team can be easily and reliably used by non-data scientists across different disciplines, greatly simplifying and speeding up the task of image analysis.
|
|
Furthermore, the scope of this reported method can be extended towards more complex analyses such as the identification of chromosome aberrations and the development of new advisory systems for object detection and classification.
|
|
“There are more trivial classifications in our lives than one might imagine. Automating such classifications by entrusting them to an AI can not only eliminate fluctuations caused by individual differences but also save many valuable research hours. Streamlining such trivial classifications make extensive image-based studies more reproducible and reliable,” says Dr. Nagaki.
|
|
Indeed, “a deep learning sorter that anyone can use,” as he puts it, could be our key to understanding various nuances of biological species.
|

