| Posted: May 19, 2009 | |
Quantum dots light up individual DNA binding proteins |
|
| (Nanowerk Spotlight) Proteins that bind to specific sites of DNA are essential to all biological functions of DNA. These DNA-binding proteins include transcription factors which modulate the process of transcription, various polymerases, nucleases which cleave DNA molecules, and histones which are involved in chromosome packaging in the cell nucleus. Developing methods to precisely determine the locations and occupancy of DNA-binding proteins is instrumental to scientists' understanding of cellular processes like gene expression and regulation. | |
| Motivated by the desire to overcome some of the inherent limitations of existing biochemical techniques for mapping protein binding sites on DNA, scientists at UCLA have now demonstrated the viability of a single molecule approach to directly visualize and map protein binding sites on DNA using fluorescent quantum dots, allowing multicolor, nanometer-resolution localization. | |
| "We harness the molecular recognition ability of DNA binding proteins to assemble nanoparticles in a controlled manner on a DNA template," Yuval Ebenstein, a post doctoral fellow at the Department of Chemistry and Biochemistry at UCLA, tells Nanowerk. "Ultimately, one can design a DNA template with any desired arrangement of unique addresses – binding sequences for specific proteins – and send quantum dots that will specifically bind to these addresses via protein-DNA interaction. These quantum dots may be designed to emit light at different colors to create unique barcodes that may be readout utilizing optical imaging." | |
| The team reported their findings in a recent edition of Nano Letters ("Lighting Up Individual DNA Binding Proteins with Quantum Dots"). | |
 |
|
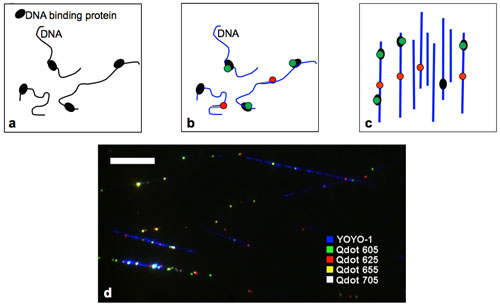
| Experimental steps for mapping DNA binding proteins. a) Crosslinking DNA-binding proteins (black) to DNA b) Staining DNA (blue), quantum dot labeling of bound proteins (green), and labeling of specific reference sequences on DNA with quantum dots (red). c) Complexes are aligned on a glass coverslip and imaged by a fluorescence microscope. Image analysis provides information on protein location. d) Pseudo color image of RNAP-biotin crosslinked to aligned DNA and bound to streptavidin quantum dots of 4 different colors (Scale-bar 10 µm). (Image: Dr. Ebenstein, UCLA) | |
| The real exciting aspect of this new technique, however, might be its potential application for genetic analysis. According to Ebenstein, with some additional development the method can be used for direct imaging of transcription factors bound to genomic DNA. | |
| In their approach, the UCLA team first crosslinks proteins to DNA using standard in vivo based ChIP protocols. | |
| "Next" explains Ebenstein, "the cross-linked complexes are labeled with quantum dot conjugated antibodies against transcription factors of interest, while the DNA is stained with an intercalating dye. Practically, four to six different transcription factors can be labeled with different quantum dots emitting in the visible spectrum and still be easily spectrally separated. Additionally, one or more spectrally distinct quantum dots could be used to tag a reference point in the genome in a sequence specific manner, using cDNA or peptide nucleic acid (PNA) probes. The quantum dot labeled protein-DNA complexes are then aligned on a glass coverslip, resulting in linear DNA molecules decorated with quantum dot labeled transcription factors." | |
| While this appears to be the first example of targeting quantum dots to DNA via molecular recognition of a DNA binding protein – in this case RNA polymerase – it is also the first single molecule level analysis of protein binding sites on a full viral genome and the researchers were able to map all known binding sites with good precision. All this is achieved in a very simple and relatively cheap way – the images can be taken with a standard, home grade, color digital camera through the microscope. | |
 |
|
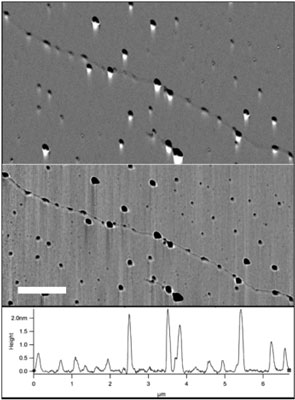
| Evaluation of RNAP and quantum dot binding on the T7-phage genome. a) Amplitude and b) phase AFM scans of the T7- DNA-RNAP-quantum dot complex. Protrusions of two typical sizes, corresponding to the RNAP and RNAP-quantum dot are visible on the DNA backbone. (Scale-bar 1 µm). c) Cross section along the DNA backbone depicting the height of the various protrusions. (Image: Dr. Ebenstein, UCLA) | |
| Ebenstein describes the potential of this technique: "We would like to be able to analyze multiple transcription factors simultaneously on long stretches of DNA. This will allow us to detect cooperative activity of different transcription factors which is very hard to do with existing techniques." | |
| Another exciting capability he mentions is the analysis of repetitive regions of the genome (accounting for about 30% of the human genome). These regions can not be distinguished one from another by conventional methods that use sequencing or DNA arrays but in the UCLA team's method can be directly localized by imaging the full genomic region of interest. | |
| In the long run, this technique may be used for diagnostics of a whole array of diseases that are caused or induced by a significant change in transcription patterns. By visually comparing the labeled genomes of healthy versus sick individuals one can identify genomic regions where differences in transcription factor binding are detected. | |
| Whereas Ebenstein and his collaborators have just demonstrated a proof of principal experiment using a viral genome and one protein, there are still many challenges for dealing with more complex genomes such as the human genome and even bacterial genomes. | |
| "These genomes are many times longer than the one we used and are likely to be fragmented," he says. "The first necessary development is to be able to mark the DNA specifically so that we know where we are on the genome and what is the orientation so that binding sites may be analyzed correctly." | |
 By
Michael
Berger
– Michael is author of three books by the Royal Society of Chemistry:
Nano-Society: Pushing the Boundaries of Technology,
Nanotechnology: The Future is Tiny, and
Nanoengineering: The Skills and Tools Making Technology Invisible
Copyright ©
Nanowerk LLC
By
Michael
Berger
– Michael is author of three books by the Royal Society of Chemistry:
Nano-Society: Pushing the Boundaries of Technology,
Nanotechnology: The Future is Tiny, and
Nanoengineering: The Skills and Tools Making Technology Invisible
Copyright ©
Nanowerk LLC
|
|
Become a Spotlight guest author! Join our large and growing group of guest contributors. Have you just published a scientific paper or have other exciting developments to share with the nanotechnology community? Here is how to publish on nanowerk.com.
