| Posted: Dec 07, 2006 | |
RNA used to control a DNA rotary nanomachine |
|
| (Nanowerk Spotlight) A large number of DNA-based nanomechanical devices have been described, controlled by a variety of methods: These include pH changes and the addition of other molecular components, such as small molecule effectors, proteins and DNA strands. The most versatile of these devices are those that are controlled by DNA strands: This versatility results because they can be addressed specifically by strands with particular sequences; these strands can be added to the solution directly, or perhaps they can result from another process ongoing within the local environment. Researchers have now shown that the state of a DNA-based nanomechanical device can be controlled by RNA strands, which means that nanomechanical devices could potentially be run from transcriptionally derived RNA molecules. | |
| Dr. Nadrian C. Seeman, professor at NYU's Chemistry Department, explains an example of a DNA nanomechanical device based on hybridization topology to Nanowerk: "Our PX-JX2 device is a robust sequence-dependent DNA nanomachine. It is controlled by the addition of DNA strands to the solution. Here is how it works:" | |
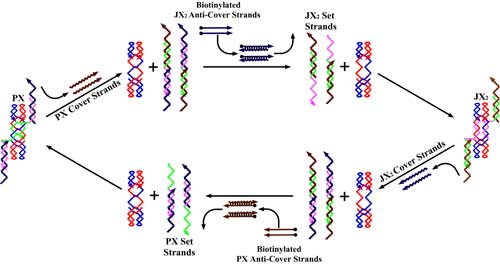
| The sequence-dependent device is based on the PX motif of DNA. The PX motif, postulated to be involved in genetic recombination, consists of two helical domains formed by four strands that flank a central dyad axis (indicated by the vertical black arrows). In (a) below, two stands are drawn in red and two in blue, where the arrowheads indicate the 3' ends of the strands. The Watson-Crick base pairing in which every nucleotide participates is indicated by the thin horizontal lines within the two double helical domains. Every possible crossover occurs between the two helical domains. The same conventions apply to the JX2 motif which lacks two crossovers in the middle. The letters A, B, C and D, along with the color coding, show that the bottom of the JX2 motif (C and D) are rotated 180° relative to the PX motif. (b) illustrates the principles of device operation. On the left is a PX molecule. The green set strands are removed by the addition of biotinylated green fuel strands (biotin indicated by black circles) in process I. The unstructured intermediate is converted to the JX2 motif by the addition of the purple set strands in process II. The JX2 molecule is converted to the unstructured intermediate by the addition of biotinylated purple fuel strands in process III. The identity of this intermediate and the one above it is indicated by the identity sign between them. The cycle is completed by the addition of green set strands in process IV, restoring the PX device. | |
 |
|
| (Image: Dr. Nadrian Seeman) | |
| The ability to control such devices with RNA will enable the machine to respond to signals generated by transcriptional logic circuits in vitro, or possibly in vivo. | |
| "The PX-JX2 device does not respond properly to RNA strands, so we have adapted the signaling method by using a cover-strand strategy" says Seeman. "This approach entails using DNA control strands that contain sequence regions to set the device to either state. An RNA strand is used to mask one of these two regions, enabling the machine to assume the state corresponding to the unmasked region." | |
| Seeman and his colleagues have previously reported the use of a pair of these devices in a larger construct that converts DNA sequences to polymer assembly instructions ("Translation of DNA Signals into Polymer Assembly Instructions"). Recently, they have developed a cassette that incorporates this device into 2D DNA arrays, thereby enabling the use of a variety of devices as individual robotic arms, potentially capable of nano-patterning and of performing nanomanufacturing operations. | |
| "A series of operations by this device can be effected by the addition of strands to the solution" Seeman explains. "However, a more general type of control would include the ability of the device to respond to signals generated within the solution. For example, these signals could report some condition within the solution, such as the RNA output of in vivo or in vitro transcriptional logic circuits. The presence of the signals would then enable the system to respond to this condition, either by active nanomechanical motion, or perhaps by recording the existence of the condition as an historical output record. To behave in this fashion, it is necessary for the PX-JX2 system to respond properly to RNA." | |
| Initially, the researchers encountered difficulties when attempting to substitute RNA directly for the DNA control strands. This was not unexpected, because the helical structures of RNA and DNA differ substantially. | |
| Seeman describes the solution to this problem: "It is well-known that RNA and DNA hybridize successfully to each other in a linear context. Thus, we have chosen to use the robust parts of the two systems, the successful response of the PX-JX2 device to conventional DNA strands and the ability of RNA to pair successfully with single-stranded DNA to yield an RNA-DNA hybrid duplex molecule. The approach adopted here is to use RNA as a 'cover strand' that masks particular oligonucleotides within a longer DNA single strand. The strategy entails using a DNA strand that contains control regions for both states. However, in the presence of the controlling RNA strands, only one of the control regions is exposed. The control strands are the same, but their coverage by different RNA strands results in different functions for them. Thus, when one portion of the strand is exposed, it binds to the device setting it to the PX state; likewise when another portion of the strand is exposed, it binds to the device setting it to the JX2 state." | |
 |
|
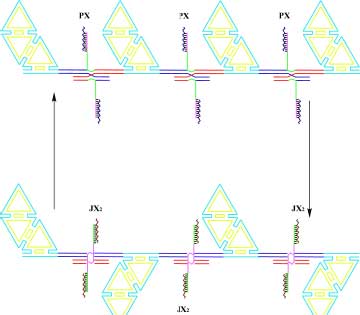
| A System to Visualize Changes of State by Atomic Force Microscopy. The top part of the diagram shows a series of half-hexagon DNA trapezoids connected by a series of PX-JX2 devices controlled by the RNA cover strand technique; the devices are in the PX state, so they are all parallel. The bottom shows that when the system is converted to the JX2 state the trapezoids form a zigzag arrangement. The system is large enough to be visualized in by the AFM. (Image: Dr. Nadrian Seeman) | |
| These findings suggest that the cover-strand approach could be used to produce translation products that reported the state of the environment of the translation machine, reflecting the results of environment-sensitive transcription. | |
| "We plan to use the devices to control the output of ribosome-like DNA devices. Systems responsive to RNA can report on the state of the solution and build molecules in response to that input" says Seeman. "This will be real hard." | |
| This recent work, titled "RNA Used to Control a DNA Rotary Nanomachine", was published in the November 30, 2006 online edition of Nano Letters. | |
 By
Michael
Berger
– Michael is author of three books by the Royal Society of Chemistry:
Nano-Society: Pushing the Boundaries of Technology,
Nanotechnology: The Future is Tiny, and
Nanoengineering: The Skills and Tools Making Technology Invisible
Copyright ©
Nanowerk LLC
By
Michael
Berger
– Michael is author of three books by the Royal Society of Chemistry:
Nano-Society: Pushing the Boundaries of Technology,
Nanotechnology: The Future is Tiny, and
Nanoengineering: The Skills and Tools Making Technology Invisible
Copyright ©
Nanowerk LLC
|
Become a Spotlight guest author! Join our large and growing group of guest contributors. Have you just published a scientific paper or have other exciting developments to share with the nanotechnology community? Here is how to publish on nanowerk.com.
