| Posted: Feb 25, 2011 | |
Functionalized nanochannels can detect single-mismatched DNA sequence |
|
| (Nanowerk Spotlight) A Single Nucleotide Polymorphism (SNP, pronounced snip) is a single nucleotide replacement in a DNA sequence – occurring when a single nucleotide (A, T, C, or G) in the genome differs – which can result in different reaction by people to pathogens and medicines. Detection of these SNPs is becoming increasingly important with the move towards more personalized healthcare by identifying inherited diseases and developing drug candidates. Researchers are therefore working hard in developing biomedical lab-on-chip sensors that allow the fast detection of SNPs in DNA using only very small samples of a patient's blood. | |
| Already, nanoscale detection techniques such as synthetic nanochannels prepared by using electron or ion beam and track-etching are being used for DNA detection by specific DNA hybridization with molecular probes immobilized on the nanochannel walls. However, the preparation of these sensors is not easy and specific functionalization at the wall surface remains a critical issues. | |
| Nanotechnology researchers at Pohang University of Science Technology (POSTECH) in South Korea have now introduced a new concept of DNA-based molecular recognition agents (MRAs) which allows detecting SNPs with very high precision and efficiency. | |
| "We have fabricated an ultrahigh-density array of functional nanochannels whose walls are covered with carboxylic acid, which served as highly sophisticated molecular recognition probes," Jin Kon Kim, a professor in the Department of Chemical Engineering, and director of National Creativity Research Initiative Program for Block Copolymer Self Assembly at POSTECH, tells Nanowerk. "In contrast to the conventional single-stranded DNA probe – which cannot differentiate the closely mismatched DNA strands with similar stability – the nanochannel system employed in our study showed high resolution to detect a single-base mismatch as well as to discriminate single-mismatched sequence at various locations." | |
| Reporting their findings in the February 6, 2011, online edition of Nano Letters ("DNA-Functionalized Nanochannels for SNP Detection"), Kim and his group introduce a simple and straightforward strategy to functionalize the nanochannels based on self-assembly of block copolymer. | |
| "We used functional groups inside the nanopores as immobilization site for MRAs" explains Kim. "The nanochannel was exploited to evaluate the potential of SNP detection. The biggest advantage of this nanochannel system is that unlike other prevalent SNP detection devices it does not require any enzyme or electrical/chemical signal. Additionally, our system detects precisely the SNP during continuous flowing. It could be employed in genetic diagnosis and separation related to molecular recognition." | |
| Since the target molecule dynamically passes through the MRA-immobilized 3D nanochannel, the frequency of the interaction between target DNA and the probe DNA increases dramatically. The result is that a subtle difference in a molecular affinity can be easily amplified and can be detected with high precision. | |
 |
|
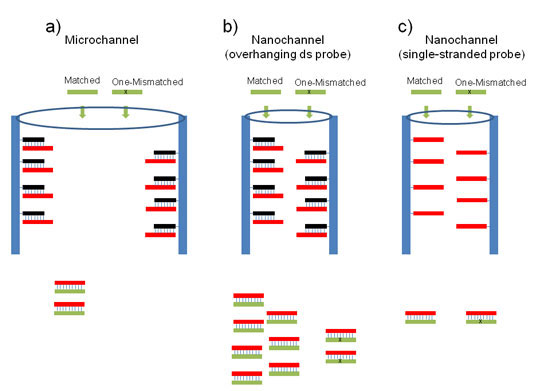
| (a) The distance between the pore walls in commonly-used microchannel typed DNA chip is too long; thus only a few perfected matched DNAs could be hybridized with the overhanging complementary DNAs located at the wall. In this case, the degree of the hybridization with one mismatched DNAs is very small (almost absence in final product). (b) The distance between the pore walls in nanochannels developed in this study are short; thus, almost all of the perfect matched DNAs could be hybridized due to the confinement effect as well as using overhanging complementary DNAs. On the other hand, only a few mismatched DNAs could be hybridized due to the thermodynamic instability. Therefore, the detection of one mismatched DNAs was achieved. (c) When a single strand probe DNA is used even in a nanochannel, the degree of the hybridization between perfectly match and single mismatched DNA is very large regardless of perfectly matched or one-mismatched DNA. There is almost no detachment from the wall, namely, the amount of the resultant ds strand in final product is extremely small. (Image: Dr. Kim, POSTECH) | |
| In conventional microarray DNA chip-based system, the thermodynamic difference between DNAs is a main factor to the detection resolution, while both thermodynamic stability and kinetic effects arising due to the special confinement of nanochannel play an important role in governing the nanochannels' high resolution (see the figure above). | |
| Furthermore, in contrast to the conventional single-stranded DNA probe which cannot differentiate the closely mismatched DNA strands with similar stability, the overhanging double-stranded DNA probe installed in the nanochannel system showed high resolution to detect a single-base mismatch as well as to discriminate single-mismatched sequences at various locations. | |
| The preparation of the ultrahigh density array (∼6 x 1011/in2) of nanochannels, decorated by functional groups in the nanopore walls, made use of block copolymer self-assembly without any post-treatment. | |
| Testing their novel nanochannel array in the lab, Kim and his team were able to distinguish a single nucleotide mismatched DNA sequence as well as the location of the mismatched DNA sequence. | |
| "Since our nanochannel system enables the precise detection of SNP during continuous flow without any enzyme or electrical/chemical signal, it could be employed in genetic diagnosis and separation related to molecular recognition" says Kim. "By detecting individual SNPs with high precision and rapidity using this nanochannel system, personalized medicine which can maximize the drug effect but minimize the side effect, could be realized in the near future." | |
| He points out that, since the properties of the nanochannels prepared by block copolymer can be easily tailored – simply by changing the end-group or chemical structure of the block copolymer – appropriately engineered nanochannels can be applied to the separation of specific materials such as chiral compounds. | |
 By
Michael
Berger
– Michael is author of three books by the Royal Society of Chemistry:
Nano-Society: Pushing the Boundaries of Technology,
Nanotechnology: The Future is Tiny, and
Nanoengineering: The Skills and Tools Making Technology Invisible
Copyright ©
Nanowerk LLC
By
Michael
Berger
– Michael is author of three books by the Royal Society of Chemistry:
Nano-Society: Pushing the Boundaries of Technology,
Nanotechnology: The Future is Tiny, and
Nanoengineering: The Skills and Tools Making Technology Invisible
Copyright ©
Nanowerk LLC
|
|
|
Become a Spotlight guest author! Join our large and growing group of guest contributors. Have you just published a scientific paper or have other exciting developments to share with the nanotechnology community? Here is how to publish on nanowerk.com. |
|
